1RTU
 
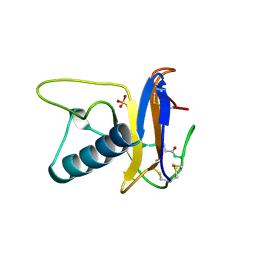 | | USTILAGO SPHAEROGENA RIBONUCLEASE U2 | | Descriptor: | RIBONUCLEASE U2, SULFATE ION | | Authors: | Noguchi, S, Satow, Y, Uchida, T, Sasaki, C, Matsuzaki, T. | | Deposit date: | 1995-05-12 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Ustilago sphaerogena ribonuclease U2 at 1.8 A resolution.
Biochemistry, 34, 1995
|
|
2E5E
 
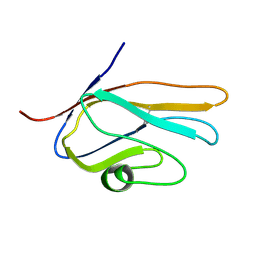 | | Solution Structure of Variable-type Domain of Human Receptor for Advanced Glycation Endproducts | | Descriptor: | Advanced glycosylation end product-specific receptor | | Authors: | Matsumoto, S, Yoshida, T, Yasumatsu, I, Yamamoto, H, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2006-12-20 | | Release date: | 2007-12-25 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Variable-type Domain of Human Receptor for Advanced Glycation Endproducts
to be published
|
|
1VA2
 
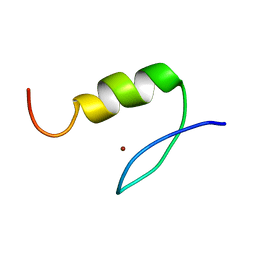 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 2) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1V8X
 
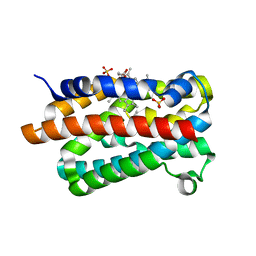 | | Crystal Structure of the Dioxygen-bound Heme Oxygenase from Corynebacterium diphtheriae | | Descriptor: | Heme oxygenase, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Unno, M, Matsui, T, Chu, G.C, Couture, M, Yoshida, T, Rousseau, D.L, Olson, J.S, Ikeda-Saito, M. | | Deposit date: | 2004-01-15 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Dioxygen-bound Heme Oxygenase from Corynebacterium diphtheriae: IMPLICATIONS FOR HEME OXYGENASE FUNCTION.
J.Biol.Chem., 279, 2004
|
|
1IPC
 
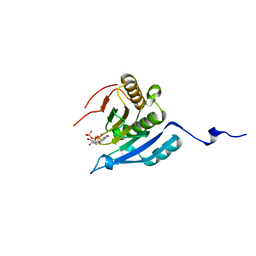 | | CRYSTAL STRUCTURE OF EUKARYOTIC INITIATION FACTOR 4E COMPLEXED WITH 7-METHYL GTP | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E | | Authors: | Tomoo, K, Shen, X, Okabe, K, Nozoe, Y, Fukuhara, S, Morino, S, Ishida, T, Taniguchi, T, Hasegawa, H, Terashima, A, Sasaki, M, Katsuya, Y, Kitamura, K, Miyoshi, H, Ishikawa, M, Miura, K. | | Deposit date: | 2001-05-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of 7-methylguanosine 5'-triphosphate (m(7)GTP)- and
P(1)-7-methylguanosine-P(3)-adenosine-5',5'-triphosphate (m(7)GpppA)-bound human full-length eukaryotic
initiation factor 4E: biological importance of the C-terminal flexible region
BIOCHEM.J., 362, 2002
|
|
1V6R
 
 | | Solution Structure of Endothelin-1 with its C-terminal Folding | | Descriptor: | Endothelin-1 | | Authors: | Takashima, H, Mimura, N, Ohkubo, T, Yoshida, T, Tamaoki, H, Kobayashi, Y. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Distributed Computing and NMR Constraint-Based High-Resolution Structure
Determination: Applied for Bioactive Peptide Endothelin-1 To Determine C-Terminal
Folding
J.Am.Chem.Soc., 126, 2004
|
|
1VA3
 
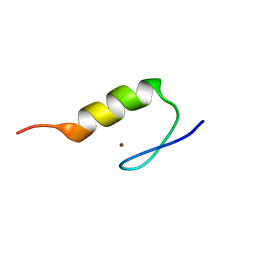 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 3) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1VA1
 
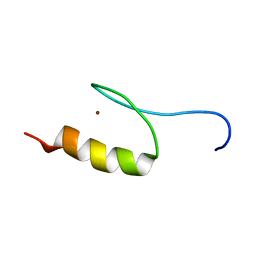 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 1) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
2N9I
 
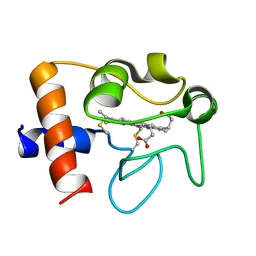 | | Solution structure of reduced human cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Imai, M, Saio, T, Kumeta, H, Uchida, T, Inagaki, F, Ishimori, K. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Investigation of the redox-dependent modulation of structure and dynamics in human cytochrome c
Biochem.Biophys.Res.Commun., 469, 2016
|
|
1WSD
 
 | | Alkaline M-protease form I crystal structure | | Descriptor: | CALCIUM ION, M-protease, SULFATE ION | | Authors: | Shirai, T, Suzuki, A, Yamane, T, Ashida, T, Kobayashi, T, Hitomi, J, Ito, S. | | Deposit date: | 2004-11-05 | | Release date: | 2004-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structure of M-protease: phylogeny aided analysis of the high-alkaline adaptation mechanism
Protein Eng., 10, 1997
|
|
1PIP
 
 | | CRYSTAL STRUCTURE OF PAPAIN-SUCCINYL-GLN-VAL-VAL-ALA-ALA-P-NITROANILIDE COMPLEX AT 1.7 ANGSTROMS RESOLUTION: NONCOVALENT BINDING MODE OF A COMMON SEQUENCE OF ENDOGENOUS THIOL PROTEASE INHIBITORS | | Descriptor: | Papain, SUCCINYL-GLN-VAL-VAL-ALA-ALA-P-NITROANILIDE | | Authors: | Yamamoto, A, Tomoo, K, Doi, M, Ohishi, H, Inoue, M, Ishida, T, Yamamoto, D, Tsuboi, S, Okamoto, H, Okada, Y. | | Deposit date: | 1992-10-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of papain-succinyl-Gln-Val-Val-Ala-Ala-p-nitroanilide complex at 1.7-A resolution: noncovalent binding mode of a common sequence of endogenous thiol protease inhibitors.
Biochemistry, 31, 1992
|
|
7FHS
 
 | | Crystal structure of DYRK1A in complex with RD0392 | | Descriptor: | (5~{Z})-5-[(3-ethoxy-4-oxidanyl-phenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, GLYCEROL | | Authors: | Kikuchi, M, Sumida, T, Hosoya, T, Kii, I, Umehara, T. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure-activity relationship for the folding intermediate-selective inhibition of DYRK1A.
Eur.J.Med.Chem., 227, 2022
|
|
5XCZ
 
 | | Structure of the cellobiohydrolase Cel6A from Phanerochaete chrysosporium in complex with cellobiose at 2.1 angstrom | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glucanase, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Tachioka, M, Nakamura, A, Ishida, T, Igarashi, K, Samejima, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-07-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a family 6 cellobiohydrolase from the basidiomycete Phanerochaete chrysosporium
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5XCY
 
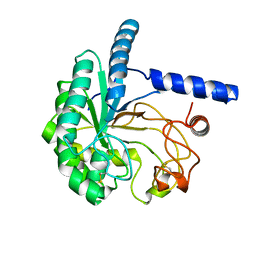 | | Structure of the cellobiohydrolase Cel6A from Phanerochaete chrysosporium at 1.2 angstrom | | Descriptor: | Glucanase | | Authors: | Tachioka, M, Nakamura, A, Ishida, T, Igarashi, K, Samejima, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Crystal structure of a family 6 cellobiohydrolase from the basidiomycete Phanerochaete chrysosporium
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
1WE1
 
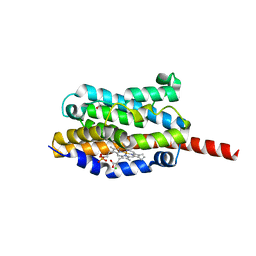 | | Crystal structure of heme oxygenase-1 from cyanobacterium Synechocystis sp. PCC6803 in complex with heme | | Descriptor: | CHLORIDE ION, Heme oxygenase 1, ISOPROPYL ALCOHOL, ... | | Authors: | Sugishima, M, Migita, C.T, Zhang, X, Yoshida, T, Fukuyama, K. | | Deposit date: | 2004-05-21 | | Release date: | 2004-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of heme oxygenase-1 from cyanobacterium Synechocystis sp. PCC 6803 in complex with heme
Eur.J.Biochem., 271, 2004
|
|
1WPK
 
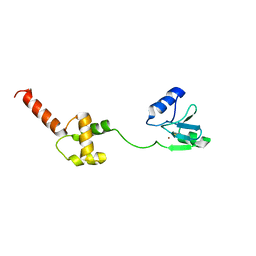 | | Methylated Form of N-terminal Transcriptional Regulator Domain of Escherichia Coli Ada Protein | | Descriptor: | ADA regulatory protein, ZINC ION | | Authors: | Takinowaki, H, Matsuda, Y, Yoshida, T, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2004-09-07 | | Release date: | 2005-09-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the methylated form of the N-terminal 16-kDa domain of Escherichia coli Ada protein
Protein Sci., 15, 2006
|
|
1WW9
 
 | | Crystal structure of the terminal oxygenase component of carbazole 1,9a-dioxygenase, a non-heme iron oxygenase system catalyzing the novel angular dioxygenation for carbazole and dioxin | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, terminal oxygenase component of carbazole | | Authors: | Nojiri, H, Ashikawa, Y, Noguchi, H, Nam, J.-W, Urata, M, Fujimoto, Z, Mizuno, H, Yoshida, T, Habe, H, Omori, T. | | Deposit date: | 2005-01-05 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the terminal oxygenase component of angular dioxygenase, carbazole 1,9a-dioxygenase
J.Mol.Biol., 351, 2005
|
|
1WKI
 
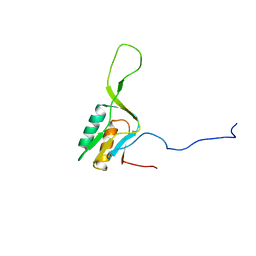 | | solution structure of ribosomal protein L16 from thermus thermophilus HB8 | | Descriptor: | LSU ribosomal protein L16P | | Authors: | Nishimura, M, Yoshida, T, Shirouzu, M, Terada, T, Kuramitsu, S, Yokoyama, S, Ohkubo, T, Kobayashi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-31 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ribosomal Protein L16 from Thermus thermophilus HB8
J.Mol.Biol., 344, 2004
|
|
1KKT
 
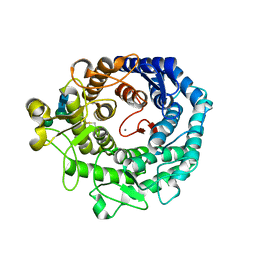 | | Structure of P. citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the ER and Golgi Class I enzymes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mannosyl-oligosaccharide alpha-1,2-mannosidase, ... | | Authors: | Lobsanov, Y.D, Vallee, F, Imberty, A, Yoshida, T, Yip, P, Herscovics, A, Howell, P.L. | | Deposit date: | 2001-12-10 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Penicillium citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the endoplasmic reticulum and Golgi class I enzymes.
J.Biol.Chem., 277, 2002
|
|
1KRF
 
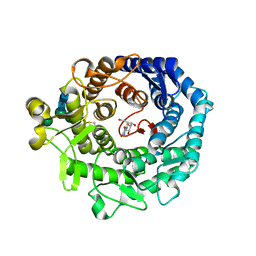 | | STRUCTURE OF P. CITRINUM ALPHA 1,2-MANNOSIDASE REVEALS THE BASIS FOR DIFFERENCES IN SPECIFICITY OF THE ER AND GOLGI CLASS I ENZYMES | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, KIFUNENSINE, ... | | Authors: | Lobsanov, Y.D, Vallee, F, Imberty, A, Yoshida, T, Yip, P, Herscovics, A, Howell, P.L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Penicillium citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the endoplasmic reticulum and Golgi class I enzymes.
J.Biol.Chem., 277, 2002
|
|
1KRE
 
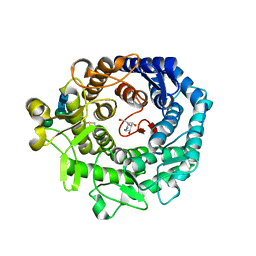 | | STRUCTURE OF P. CITRINUM ALPHA 1,2-MANNOSIDASE REVEALS THE BASIS FOR DIFFERENCES IN SPECIFICITY OF THE ER AND GOLGI CLASS I ENZYMES | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lobsanov, Y.D, Vallee, F, Imberty, A, Yoshida, T, Yip, P, Herscovics, A, Howell, P.L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Penicillium citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the endoplasmic reticulum and Golgi class I enzymes.
J.Biol.Chem., 277, 2002
|
|
1Y1C
 
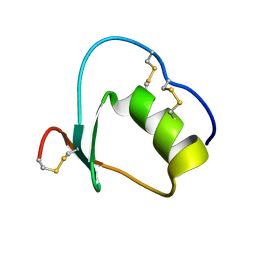 | | Solution structure of Anemonia elastase inhibitor analogue | | Descriptor: | Elastase inhibitor | | Authors: | Hemmi, H, Kumazaki, T, Yoshizawa-Kumagaye, K, Nishiuchi, Y, Yoshida, T, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2004-11-18 | | Release date: | 2005-07-19 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Study of an Anemonia Elastase Inhibitor, a "Nonclassical" Kazal-Type Inhibitor from Anemonia sulcata
Biochemistry, 44, 2005
|
|
1Y1B
 
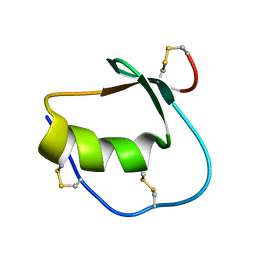 | | Solution structure of Anemonia elastase inhibitor | | Descriptor: | Elastase inhibitor | | Authors: | Hemmi, H, Kumazaki, T, Yoshizawa-Kumagaye, K, Nishiuchi, Y, Yoshida, T, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2004-11-18 | | Release date: | 2005-07-19 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Study of an Anemonia Elastase Inhibitor, a "Nonclassical" Kazal-Type Inhibitor from Anemonia sulcata
Biochemistry, 44, 2005
|
|
1CYC
 
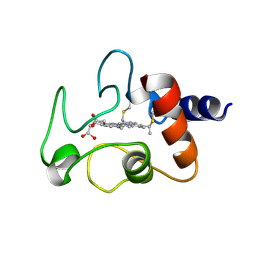 | | THE CRYSTAL STRUCTURE OF BONITO (KATSUO) FERROCYTOCHROME C AT 2.3 ANGSTROMS RESOLUTION. II. STRUCTURE AND FUNCTION | | Descriptor: | FERROCYTOCHROME C, HEME C | | Authors: | Tanaka, N, Yamane, T, Tsukihara, T, Ashida, T, Kakudo, M. | | Deposit date: | 1976-08-01 | | Release date: | 1976-10-06 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of bonito (katsuo) ferrocytochrome c at 2.3 A resolution. II. Structure and function.
J.Biochem.(Tokyo), 77, 1975
|
|
1MPY
 
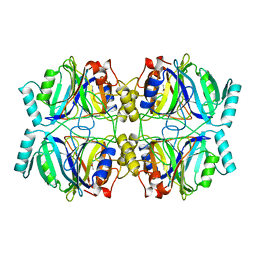 | | STRUCTURE OF CATECHOL 2,3-DIOXYGENASE (METAPYROCATECHASE) FROM PSEUDOMONAS PUTIDA MT-2 | | Descriptor: | ACETONE, CATECHOL 2,3-DIOXYGENASE, FE (II) ION | | Authors: | Kita, A, Kita, S, Fujisawa, I, Inaka, K, Ishida, T, Horiike, K, Nozaki, M, Miki, K. | | Deposit date: | 1998-10-20 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An archetypical extradiol-cleaving catecholic dioxygenase: the crystal structure of catechol 2,3-dioxygenase (metapyrocatechase) from Ppseudomonas putida mt-2.
Structure Fold.Des., 7, 1999
|
|
