5WVR
 
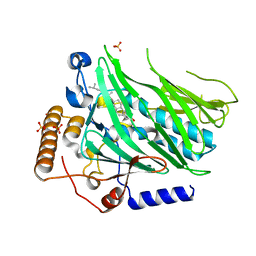 | | Crystal structure of Osh1 ORD domain in complex with cholesterol | | Descriptor: | CHOLESTEROL, KLLA0C04147p, SULFATE ION | | Authors: | Im, Y.J, Manik, M.K, Yang, H.S, Tong, J.S. | | Deposit date: | 2016-12-28 | | Release date: | 2017-05-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Yeast OSBP-Related Protein Osh1 Reveals Key Determinants for Lipid Transport and Protein Targeting at the Nucleus-Vacuole Junction
Structure, 25, 2017
|
|
5H2D
 
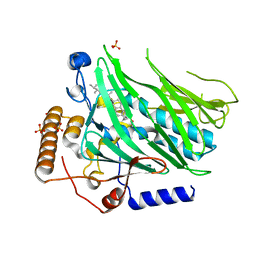 | | Crystal structure of Osh1 ORD domain in complex with ergosterol | | Descriptor: | ERGOSTEROL, KLLA0C04147p, SULFATE ION | | Authors: | Im, Y.J, Manik, M.K, Yang, H.S, Tong, J.S. | | Deposit date: | 2016-10-14 | | Release date: | 2017-05-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Yeast OSBP-Related Protein Osh1 Reveals Key Determinants for Lipid Transport and Protein Targeting at the Nucleus-Vacuole Junction
Structure, 25, 2017
|
|
5H2A
 
 | |
5H28
 
 | |
5H2C
 
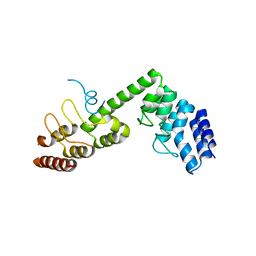 | | Crystal structure of Saccharomyces cerevisiae Osh1 ANK - Nvj1 | | Descriptor: | Nucleus-vacuole junction protein 1, Oxysterol-binding protein homolog 1 | | Authors: | Im, Y.J, Manik, M.K, Yang, H.S, Tong, J.S. | | Deposit date: | 2016-10-14 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.508 Å) | | Cite: | Structure of Yeast OSBP-Related Protein Osh1 Reveals Key Determinants for Lipid Transport and Protein Targeting at the Nucleus-Vacuole Junction
Structure, 25, 2017
|
|
5Y2T
 
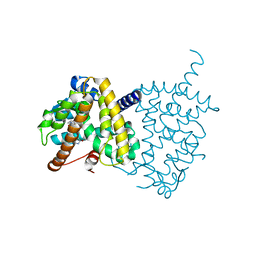 | | Structure of PPARgamma ligand binding domain - lobeglitazone complex | | Descriptor: | (5S)-5-[[4-[2-[[6-(4-methoxyphenoxy)pyrimidin-4-yl]-methyl-amino]ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Im, Y.J, Lee, M. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of PPAR gamma complexed with lobeglitazone and pioglitazone reveal key determinants for the recognition of antidiabetic drugs
Sci Rep, 7, 2017
|
|
5Y2O
 
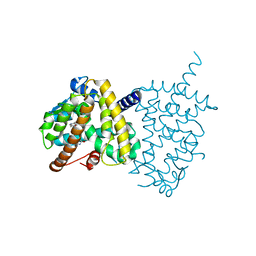 | | Structure of PPARgamma ligand binding domain-pioglitazone complex | | Descriptor: | (5S)-5-[[4-[2-(5-ethylpyridin-2-yl)ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Im, Y.J, Lee, M. | | Deposit date: | 2017-07-26 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structures of PPAR gamma complexed with lobeglitazone and pioglitazone reveal key determinants for the recognition of antidiabetic drugs
Sci Rep, 7, 2017
|
|
5YQJ
 
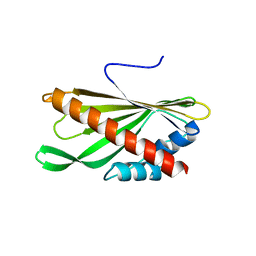 | | Crystal structure of the first StARkin domain of Lam4 | | Descriptor: | Membrane-anchored lipid-binding protein LAM4 | | Authors: | Im, Y.J, Tong, J.S. | | Deposit date: | 2017-11-06 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5AYZ
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE PRODUCT NICOTINATE MONONUCLEOTIDE | | Descriptor: | NICOTINATE MONONUCLEOTIDE, Nicotinate-nucleotide pyrophosphorylase [carboxylating] | | Authors: | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
5AYY
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE REACTANT QUINOLINATE | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating], QUINOLINIC ACID | | Authors: | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
6WCW
 
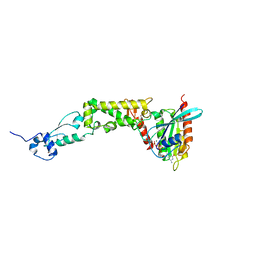 | | Structure of human Rubicon RH domain in complex with GTP-bound Rab7 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-7a, ... | | Authors: | Bhargava, H.K, Byck, J.M, Farrell, D.P, Anishchenko, I, DiMaio, F, Im, Y.J, Hurley, J.H. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for autophagy inhibition by the human Rubicon-Rab7 complex.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6L1D
 
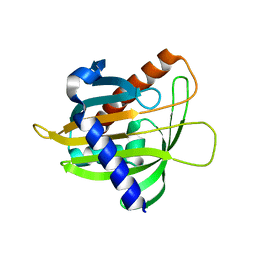 | |
6L1M
 
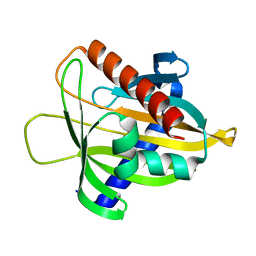 | |
3G9H
 
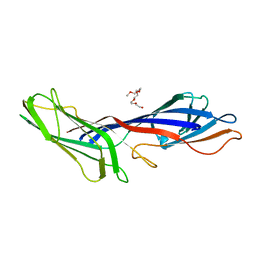 | | Crystal structure of the C-terminal mu homology domain of Syp1 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Suppressor of yeast profilin deletion | | Authors: | Reider, A, Barker, S, Mishra, S, Im, Y.J, Maldonado-Baez, L, Hurley, J, Traub, L, Wendland, B. | | Deposit date: | 2009-02-13 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation.
Embo J., 28, 2009
|
|
3G9G
 
 | | Crystal Structure of the N-terminal EFC/F-BAR domain of Syp1 | | Descriptor: | Suppressor of yeast profilin deletion | | Authors: | Reider, A, Barker, S, Mishra, S, Im, Y.J, Maldonado-Baez, L, Hurley, J, Traub, L, Wendland, B. | | Deposit date: | 2009-02-13 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation.
Embo J., 28, 2009
|
|
2FJK
 
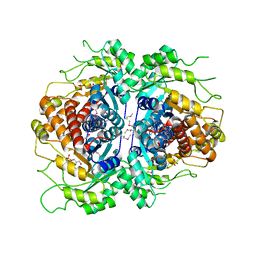 | | Crystal structure of Fructose-1,6-Bisphosphate Aldolase in Thermus caldophilus | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase | | Authors: | Lee, J.H, Im, Y.J, Rho, S.-H, Kim, M.-K, Kang, G.B, Eom, S.H. | | Deposit date: | 2006-01-03 | | Release date: | 2006-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stereoselectivity of fructose-1,6-bisphosphate aldolase in Thermus caldophilus
Biochem.Biophys.Res.Commun., 347, 2006
|
|
2IE8
 
 | |
8YJC
 
 | | Structure of Vibrio vulnificus MARTX cysteine protease domain C3727A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, INOSITOL HEXAKISPHOSPHATE, Multifunctional autoprocessing repeat-in-toxin (MARTX), ... | | Authors: | Chen, L, Khan, H, Tan, L, Li, X, Zhang, G, Im, Y.J. | | Deposit date: | 2024-03-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of the activation of MARTX cysteine protease domain from Vibrio vulnificus.
Plos One, 19, 2024
|
|
8YJA
 
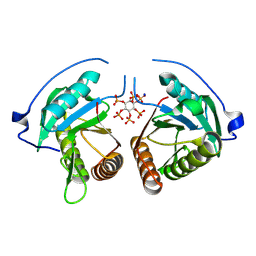 | | Structure of Vibrio vulnificus MARTX cysteine protease domain lacking beta-flap | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, MARTX cysteine protease domain, SODIUM ION | | Authors: | Chen, L, Khan, H, Tan, L, Li, X, Zhang, G, Im, Y.J. | | Deposit date: | 2024-03-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the activation of MARTX cysteine protease domain from Vibrio vulnificus.
Plos One, 19, 2024
|
|
1G4B
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
3P9H
 
 | |
4N9N
 
 | |
3P9G
 
 | |
1G4A
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
1XNG
 
 | | Crystal Structure of NH3-dependent NAD+ synthetase from Helicobacter pylori | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, ... | | Authors: | Kang, G.B, Kim, Y.S, Im, Y.J, Rho, S.H, Lee, J.H, Eom, S.H. | | Deposit date: | 2004-10-05 | | Release date: | 2005-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of NH3-dependent NAD+ synthetase from Helicobacter pylori
Proteins, 58, 2005
|
|
