7O3C
 
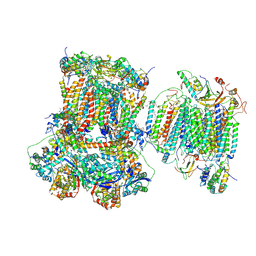 | | Murine supercomplex CIII2CIV in the mature unlocked conformation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and assembly of the mammalian mitochondrial supercomplex CIII 2 CIV.
Nature, 598, 2021
|
|
1JYQ
 
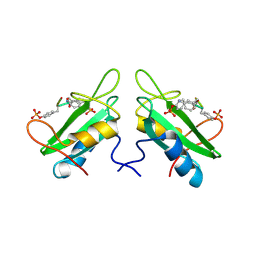 | | Xray Structure of Grb2 SH2 Domain Complexed with a Highly Affine Phospho Peptide | | Descriptor: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, mAZ-pY-(alpha Me)pY-N-NH2 peptide inhibitor | | Authors: | Nioche, P, Liu, W.-Q, Broutin, I, Charbonnier, F, Latreille, M.-T, Vidal, M, Roques, B, Garbay, C, Ducruix, A. | | Deposit date: | 2001-09-13 | | Release date: | 2002-03-13 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the SH2 domain of Grb2: highlight on the binding of a new high-affinity inhibitor.
J.Mol.Biol., 315, 2002
|
|
7O37
 
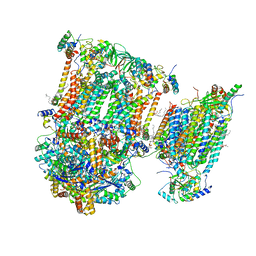 | | Murine supercomplex CIII2CIV in the assembled locked conformation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and assembly of the mammalian mitochondrial supercomplex CIII 2 CIV.
Nature, 598, 2021
|
|
7O3H
 
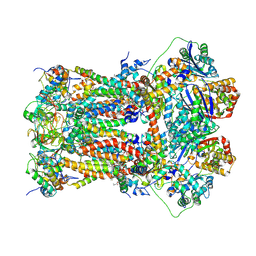 | | Murine CIII2 focus-refined from supercomplex CICIII2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure and assembly of the mammalian mitochondrial supercomplex CIII 2 CIV.
Nature, 598, 2021
|
|
6CI2
 
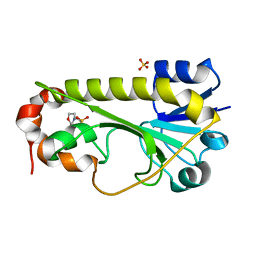 | | Crystal structure of the formyltransferase PseJ from Anoxybacillus kamchatkensis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, formyltransferase PseJ | | Authors: | Reimer, J.M, Jiang, J, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Insight into a Novel Formyltransferase and Evolution to a Nonribosomal Peptide Synthetase Tailoring Domain.
ACS Chem. Biol., 13, 2018
|
|
6TJD
 
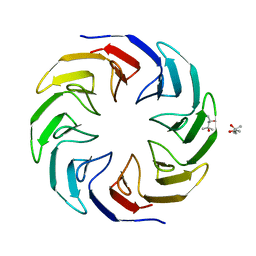 | | Crystal structure of the computationally designed Cake4 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cake4 | | Authors: | Laier, I, Mylemans, B, Noguchi, H, Voet, A.R.D. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
6THG
 
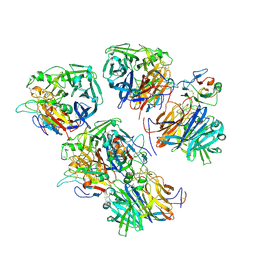 | | Cedar Virus attachment glycoprotein (G) in complex with human ephrin-B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein, ... | | Authors: | Pryce, R, Rissanen, I, Harlos, K, Bowden, T. | | Deposit date: | 2019-11-20 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.074 Å) | | Cite: | A key region of molecular specificity orchestrates unique ephrin-B1 utilization by Cedar virus.
Life Sci Alliance, 3, 2020
|
|
6TJB
 
 | | Crystal structure of the computationally designed Cake2 protein | | Descriptor: | Cake2, GLYCEROL | | Authors: | Laier, I, Mylemans, B, Noguchi, H, Voet, A.R.D. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
6TJI
 
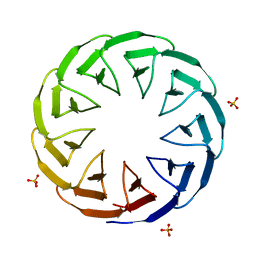 | | Crystal structure of the computationally designed Cake10 protein | | Descriptor: | Cake10, PHOSPHATE ION | | Authors: | Laier, I, Mylemans, B, Voet, A.R.D, Noguchi, H. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
1K5H
 
 | | 1-deoxy-D-xylulose-5-phosphate reductoisomerase | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate reductoisomerase | | Authors: | Reuter, K, Sanderbrand, S, Jomaa, H, Wiesner, J, Steinbrecher, I, Beck, E, Hintz, M, Klebe, G, Stubbs, M.T. | | Deposit date: | 2001-10-10 | | Release date: | 2002-02-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 1-deoxy-D-xylulose-5-phosphate reductoisomerase, a crucial enzyme in the non-mevalonate pathway of isoprenoid biosynthesis.
J.Biol.Chem., 277, 2002
|
|
1K7E
 
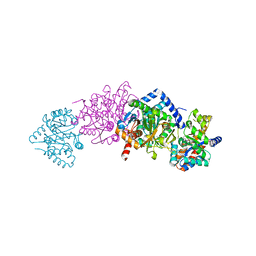 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID | | Descriptor: | N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-19 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of a new class of allosteric effectors complexed to tryptophan synthase.
J.Biol.Chem., 277, 2002
|
|
6D36
 
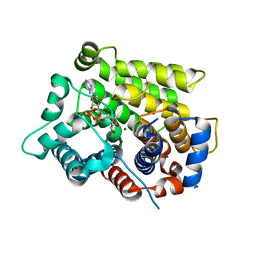 | | Structure of human ARH3 bound to ADP-ribose and magnesium | | Descriptor: | MAGNESIUM ION, Poly(ADP-ribose) glycohydrolase ARH3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Pourfarjam, Y, Ventura, J, Kurinov, I, Kim, I.K. | | Deposit date: | 2018-04-14 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human ADP-ribosyl-acceptor hydrolase 3 bound to ADP-ribose reveals a conformational switch that enables specific substrate recognition.
J.Biol.Chem., 293, 2018
|
|
6CN1
 
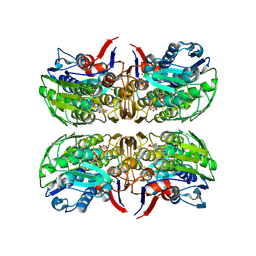 | | 2.75 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Pseudomonas putida in Complex with Uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and Magnesium | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Pseudomonas putida in Complex with Uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and Magnesium.
To Be Published
|
|
6D3A
 
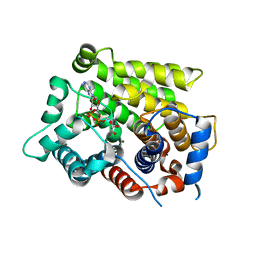 | | Structure of human ARH3 D314E bound to ADP-ribose and magnesium | | Descriptor: | MAGNESIUM ION, Poly(ADP-ribose) glycohydrolase ARH3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Pourfarjam, Y, Ventura, J, Kurinov, I, Kim, I.K. | | Deposit date: | 2018-04-15 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.60001016 Å) | | Cite: | Structure of human ADP-ribosyl-acceptor hydrolase 3 bound to ADP-ribose reveals a conformational switch that enables specific substrate recognition.
J.Biol.Chem., 293, 2018
|
|
7O01
 
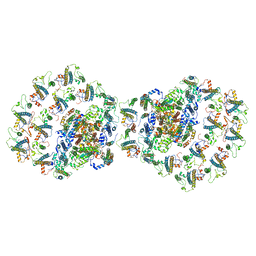 | |
7NWT
 
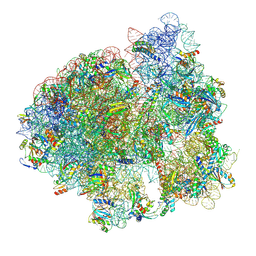 | | Initiated 70S ribosome in complex with 2A protein from encephalomyocarditis virus (EMCV) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Hill, C.H, Napthine, S, Pekarek, L, Kibe, A, Firth, A.E, Graham, S.C, Caliskan, N, Brierley, I. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural and molecular basis for Cardiovirus 2A protein as a viral gene expression switch
Nat Commun, 12, 2021
|
|
6T6Q
 
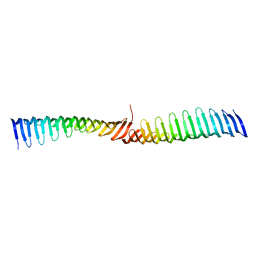 | | Crystal structure of Toxoplasma gondii Morn1 (extended conformation). | | Descriptor: | Membrane occupation and recognition nexus protein MORN1 | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
6CQF
 
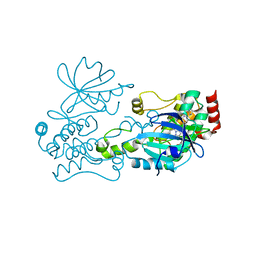 | | Crystal structure of HPK1 in complex an inhibitor G1858 | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 1, N-{2-(3,3-difluoropyrrolidin-1-yl)-6-[(3R)-pyrrolidin-3-yl]pyrimidin-4-yl}-1-(propan-2-yl)-1H-pyrazolo[4,3-c]pyridin-6-amine | | Authors: | Wu, P, Lehoux, I, Mortara, K, Franke, Y, Chan, B.K, Wang, W. | | Deposit date: | 2018-03-15 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Hematopoietic Progenitor Kinase-1 Structure in a Domain-Swapped Dimer.
Structure, 27, 2019
|
|
1K8D
 
 | | crystal structure of the non-classical MHC class Ib Qa-2 complexed with a self peptide | | Descriptor: | 60S RIBOSOMAL PROTEIN, BETA-2-MICROGLOBULIN, QA-2 antigen | | Authors: | He, X, Tabaczewski, P, Ho, J, Stroynowski, I, Garcia, K.C. | | Deposit date: | 2001-10-23 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Promiscuous antigen presentation by the nonclassical MHC Ib Qa-2 is enabled by a shallow, hydrophobic groove and self-stabilized peptide conformation.
Structure, 9, 2001
|
|
1K8Z
 
 | | CRYSTAL STRUCTURE OF THE TRYPTOPHAN SYNTHASE BETA-SER178PRO MUTANT COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID | | Descriptor: | N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-26 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the beta Ser178--> Pro mutant of tryptophan synthase. A "knock-out" allosteric enzyme.
J.Biol.Chem., 277, 2002
|
|
1K44
 
 | |
1K3O
 
 | | Crystal Structure Analysis of apo Glutathione S-Transferase | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1 | | Authors: | Le Trong, I, Stenkamp, R.E, Ibarra, C, Atkins, W.M, Adman, E.T. | | Deposit date: | 2001-10-03 | | Release date: | 2002-10-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.3-A resolution structure of human glutathione S-transferase with S-hexyl glutathione bound reveals possible extended ligandin binding site.
Proteins, 48, 2002
|
|
6D0G
 
 | | 1.78 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii | | Descriptor: | BROMIDE ION, MANGANESE (II) ION, Pirin family protein | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Kiryukhina, O, Endres, M, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-10 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 1.78 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii.
To be Published
|
|
1K7X
 
 | | CRYSTAL STRUCTURE OF THE BETA-SER178PRO MUTANT OF TRYPTOPHAN SYNTHASE | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-22 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the beta Ser178--> Pro mutant of tryptophan synthase. A "knock-out" allosteric enzyme.
J.Biol.Chem., 277, 2002
|
|
7ODL
 
 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C1 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2021-04-29 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
