5NNP
 
 | | Structure of Naa15/Naa10 bound to HypK-THB | | Descriptor: | CARBOXYMETHYL COENZYME *A, GLYCEROL, N-terminal acetyltransferase-like protein, ... | | Authors: | Weyer, F.A, Gumiero, A, Kopp, J, Sinning, I. | | Deposit date: | 2017-04-10 | | Release date: | 2017-06-14 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structural basis of HypK regulating N-terminal acetylation by the NatA complex.
Nat Commun, 8, 2017
|
|
6O3G
 
 | |
6NQP
 
 | | Crystal structure of fast switching M159T mutant of fluorescent protein Dronpa (Dronpa2), Y63(2,3-F2Y) | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Lin, C.-Y, Romei, M.G, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2019-01-21 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Electrostatic control of photoisomerization pathways in proteins.
Science, 367, 2020
|
|
1F90
 
 | | FAB FRAGMENT OF MONOCLONAL ANTIBODY (LNKB-2) AGAINST HUMAN INTERLEUKIN-2 IN COMPLEX WITH ANTIGENIC PEPTIDE | | Descriptor: | ANTIGENIC NONAPEPTIDE, FAB FRAGMENT OF MONOCLONAL ANTIBODY | | Authors: | Afonin, P.V, Fokin, A.V, Tsigannik, I.N, Mikhailova, I.Y, Onoprienko, L.V, Mikhaleva, I.I, Ivanov, V.T, Mareeva, T.Y, Nesmeyanov, V.A, Li, N, Duax, W.L, Pletnev, V.Z. | | Deposit date: | 2000-07-06 | | Release date: | 2001-07-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an anti-interleukin-2 monoclonal antibody Fab complexed with an antigenic nonapeptide.
Protein Sci., 10, 2001
|
|
6QKD
 
 | | CRYSTAL STRUCTURE OF vhh-based FAB-fragment of antibody BCD-085 | | Descriptor: | CHLORIDE ION, FAB HEAVY CHAIN, FAB LIGHT CHAIN, ... | | Authors: | Kostareva, O.S, Kolyadenko, I.A, Ulitin, A.B, Ekimova, V.M, Evdokimov, S.R, Garber, M.B, Tishchenko, T.V, Gabdulkhakov, A.G. | | Deposit date: | 2019-01-29 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fab Fragment of VHH-Based Antibody Netakimab: Crystal Structure and Modeling Interaction with Cytokine IL-17A
Crystals, 2019
|
|
7QD0
 
 | | Structure of the orange carotenoid protein from Planktothrix agardhii binding echinenone in the C2 space group | | Descriptor: | ACETATE ION, ARGININE, GLYCEROL, ... | | Authors: | Andreeva, E.A, Hartmann, E, Schlichting, I, Colletier, J.-P. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function-dynamics relationships in the peculiar Planktothrix PCC7805 OCP1: Impact of his-tagging and carotenoid type.
Biochim Biophys Acta Bioenerg, 1863, 2022
|
|
8FG5
 
 | | Apo mouse acidic mammalian chitinase, catalytic domain at 100 K | | Descriptor: | Acidic mammalian chitinase, MAGNESIUM ION | | Authors: | Diaz, R.E, Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2022-12-12 | | Release date: | 2023-03-01 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase.
Biorxiv, 2024
|
|
1MWA
 
 | | 2C/H-2KBM3/DEV8 ALLOGENEIC COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2C T CELL RECEPTOR ALPHA CHAIN, ... | | Authors: | Luz, J.G, Huang, M.D, Garcia, K.C, Rudolph, M.G, Teyton, L, Wilson, I.A. | | Deposit date: | 2002-09-27 | | Release date: | 2002-11-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural comparison of allogeneic and syngeneic T cell receptor-peptide-major histocompatibility
complex complexes: a buried alloreactive mutation subtly alters peptide presentation substantially
increasing V(beta) Interactions.
J.EXP.MED., 195, 2002
|
|
7QD2
 
 | | Structure of the orange carotenoid protein from Planktothrix agardhii binding canthaxanthin in the P21 space group | | Descriptor: | ACETATE ION, GLYCEROL, Orange carotenoid-binding protein, ... | | Authors: | Andreeva, E.A, Hartmann, E, Schlichting, I, Colletier, J.-P. | | Deposit date: | 2021-11-26 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-function-dynamics relationships in the peculiar Planktothrix PCC7805 OCP1: Impact of his-tagging and carotenoid type.
Biochim Biophys Acta Bioenerg, 1863, 2022
|
|
7T3J
 
 | | Cryo-EM structure of Csy-AcrIF24 | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
6Q44
 
 | | Est3 telomerase subunit in the yeast Hansenula polymorpha | | Descriptor: | Uncharacterized protein | | Authors: | Mantsyzov, A.B, Mariasina, S.S, Petrova, O.A, Efimov, S.V, Dontsova, O.A, Polshakov, V.I. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-25 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Insights into the structure and function of Est3 from the Hansenula polymorpha telomerase.
Sci Rep, 10, 2020
|
|
7TAX
 
 | | Cryo-EM structure of the Csy-AcrIF24-promoter DNA complex | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-21 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
3MXB
 
 | | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*CP*TP*GP*GP*CP*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*GP*AP*GP*AP*AP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*TP*TP*CP*TP*CP*AP*GP*GP*TP*AP*CP*CP*TP*CP*AP*GP*CP*CP*AP*GP*A)-3'), ... | | Authors: | Munoz, I.G, Prieto, J, Subramanian, S, Coloma, J, Montoya, G. | | Deposit date: | 2010-05-07 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus.
Nucleic Acids Res., 39, 2011
|
|
6YC2
 
 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric form at room temperature, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ALANINE, EICOSANE, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
6YFE
 
 | | Virus-like particle of Beihai levi-like virus 19 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.794 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YFO
 
 | | Virus-like particle of bacteriophage GQ-907 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.484 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
1KJV
 
 | | TAP-B-associated rat MHC class I molecule | | Descriptor: | Mature alpha chain of major histocompatibility complex class I antigen (HEAVY CHAIN), SULFATE ION, beta-2-microglobulin, ... | | Authors: | Rudolph, M.G, Stevens, J, Speir, J.A, Trowsdale, J, Butcher, G.W, Joly, E, Wilson, I.A. | | Deposit date: | 2001-12-05 | | Release date: | 2002-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of two rat MHC class Ia (RT1-A) molecules that are associated differentially
with peptide transporter alleles TAP-A and TAP-B.
J.Mol.Biol., 324, 2002
|
|
4RPI
 
 | | Crystal Structure of Nicotinate Mononucleotide Adenylyltransferase from Mycobacterium tuberculosis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, nicotinate-nucleotide adenylyltransferase | | Authors: | Rodionova, I, Zuccola, H, Sorci, L, Aleshin, A.E, Kazanov, M, Sergienko, E, Rubin, E, Locher, C, Osterman, A. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.417 Å) | | Cite: | Mycobacterial nicotinate mononucleotide adenylyltransferase: structure, mechanism, and implications for drug discovery.
J. Biol. Chem., 290, 2015
|
|
8QTD
 
 | | Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab | | Descriptor: | Spike glycoprotein,Fibritin, XBB-7 fab heavy chain, XBB-7 fab light chain | | Authors: | Ren, J, Duyvesteyn, H.M.E, Stuart, D.I. | | Deposit date: | 2023-10-12 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8QRF
 
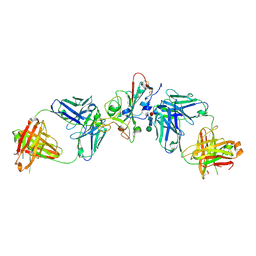 | | SARS-CoV-2 delta RBD complexed with XBB-6 and beta-49 Fabs | | Descriptor: | Beta-49 heavy chain, Beta-49 light chain, Spike protein S1, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
5NDV
 
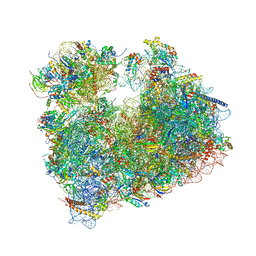 | | Crystal structure of Paromomycin bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Prokhorova, I, Djumagulov, M, Urzhumtsev, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2017-03-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Aminoglycoside interactions and impacts on the eukaryotic ribosome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3N02
 
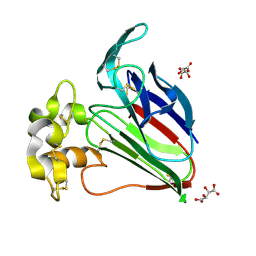 | | Thaumatic crystals grown in loops/micromounts | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Mathes, I.I. | | Deposit date: | 2010-05-13 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diffraction study of protein crystals grown in cryoloops and micromounts.
J.Appl.Crystallogr., 43, 2010
|
|
5NUN
 
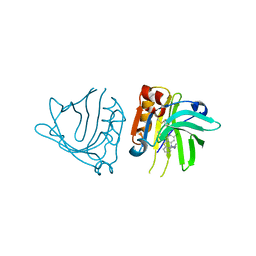 | | Engineered beta-lactoglobulin: variant F105L-L39A-M107F in complex with chlorpromazine (LG-LAF-CLP) | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Beta-lactoglobulin | | Authors: | Loch, J.I, Bonarek, P, Towrzydlo, M, Lazinskia, I, Szydlowska, J, Lewinski, K. | | Deposit date: | 2017-04-30 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The engineered beta-lactoglobulin with complementarity to the chlorpromazine chiral conformers.
Int. J. Biol. Macromol., 114, 2018
|
|
8BBN
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.2-10 and EY6A Fabs | | Descriptor: | BA.2-10 heavy chain, BA.2-10 light chain, EY6A Heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
8BCZ
 
 | | SARS-CoV-2 Delta-RBD complexed with Fabs BA.2-36, BA.2-23, EY6A and COVOX-45 | | Descriptor: | BA.2-23 heavy chain, BA.2-23 light chain, BA.2-36 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-17 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
