2V5V
 
 | | W57E Flavodoxin from Anabaena | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, MAGNESIUM ION | | Authors: | Herguedas, B, Martinez-Julvez, M, Perez-Dorado, I, Goni, G, Medina, M, Hermoso, J.A. | | Deposit date: | 2007-07-10 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Tuning of the Fmn Binding and Oxido-Reduction Properties by Neighboring Side Chains in Anabaena Flavodoxin.
Arch.Biochem.Biophys., 467, 2007
|
|
3NVH
 
 | |
1F6Z
 
 | |
1F6Y
 
 | |
1F78
 
 | |
1YMW
 
 | | The study of reductive unfolding pathways of RNase A (Y92G mutant) | | Descriptor: | Ribonuclease pancreatic | | Authors: | Xu, G, Narayan, M, Kurinov, I, Ripoll, D.R, Welker, E, Khalili, M, Ealick, S.E, Scheraga, H.A. | | Deposit date: | 2005-01-21 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A localized specific interaction alters the unfolding pathways of structural homologues.
J.Am.Chem.Soc., 128, 2006
|
|
1F79
 
 | |
132L
 
 | |
3NX7
 
 | | Crystal structure of the catalytic domain of human MMP12 complexed with the inhibitor N-Hydroxy-2-(N-(2-hydroxyethyl)4-methoxyphenylsulfonamido)acetamide | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-hydroxy-N~2~-(2-hydroxyethyl)-N~2~-[(4-methoxyphenyl)sulfonyl]glycinamide, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Giachetti, A, Loconte, M, Luchinat, C, Maletta, M, Nativi, C, Yeo, K.J. | | Deposit date: | 2010-07-13 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring the subtleties of drug-receptor interactions: the case of matrix metalloproteinases
J.Am.Chem.Soc., 129, 2007
|
|
2VM5
 
 | | HUMAN BIR2 DOMAIN OF BACULOVIRAL INHIBITOR OF APOPTOSIS REPEAT- CONTAINING 1 (BIRC1) | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 1, GLYCEROL, ZINC ION | | Authors: | Herman, M.D, Welin, M, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-23 | | Release date: | 2008-01-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Bir Domains from Human Naip and Ciap2.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2VID
 
 | | Serine protease SplB from Staphylococcus aureus at 1.8A resolution | | Descriptor: | SERINE PROTEASE SPLB | | Authors: | Dubin, G, Stec-Niemczyk, J, Kisielewska, M, Pustelny, K, Popowicz, G.M, Bista, M, Kantyka, T, Boulware, K.T, Stennicke, H.R, Czarna, A, Phopaisarn, M, Daugherty, P.S, Thogersen, I.B, Enghild, J.J, Thornberry, N, Dubin, A, Potempa, J. | | Deposit date: | 2007-11-30 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enzymatic Activity of the Staphylococcus Aureus Splb Serine Protease is Induced by Substrates Containing the Sequence Trp-Glu-Leu-Gln.
J.Mol.Biol., 379, 2008
|
|
4QFY
 
 | | Crystal structure of the tetrameric dGTP/dCTP-bound SAMHD1 (RN206) mutant catalytic core | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Koharudin, L.M.I, Wu, Y, DeLucia, M, Mehrens, J, Gronenborn, A.M, Ahn, J. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Allosteric Activation of Sterile alpha Motif and Histidine-Aspartate Domain-containing Protein 1 (SAMHD1) by Nucleoside Triphosphates.
J.Biol.Chem., 289, 2014
|
|
4PWP
 
 | | Crystal structure of DsbA from the Gram positive bacterium Corynebacterium diphtheriae | | Descriptor: | DsbA, GLYCEROL | | Authors: | Um, S.H, Kim, J.S, Jiao, L, Yoon, B.Y, Jo, I, Ha, N.C. | | Deposit date: | 2014-03-21 | | Release date: | 2015-03-25 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Crystal structure and biochemical characterization of DsbA from the Gram positive bacterium Corynebacterium diphtheriae
To be Published
|
|
4PXD
 
 | | The crystal structure of EcAAH in complex with allantoate | | Descriptor: | ALLANTOATE ION, Allantoate amidohydrolase, MANGANESE (II) ION | | Authors: | Shin, I, Rhee, S. | | Deposit date: | 2014-03-23 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the substrate specificity of (s)-ureidoglycolate amidohydrolase and its comparison with allantoate amidohydrolase.
J.Mol.Biol., 426, 2014
|
|
2VKN
 
 | | YEAST SHO1 SH3 DOMAIN COMPLEXED WITH A PEPTIDE FROM PBS2 | | Descriptor: | MAP KINASE KINASE PBS2, PROTEIN SSU81, SULFATE ION | | Authors: | Kursula, P, Kursula, I, Song, Y.H, Paraskevopoulos, K, Wilmanns, M. | | Deposit date: | 2007-12-20 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Genomics of Yeast SH3 Domains
To be Published
|
|
1YOW
 
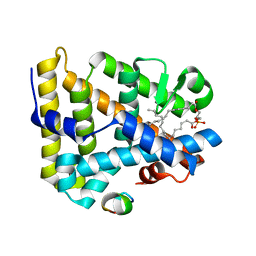 | | human Steroidogenic Factor 1 LBD with bound Co-factor Peptide | | Descriptor: | PHOSPHATIDYL ETHANOL, Steroidogenic factor 1, TIF2 peptide | | Authors: | Krylova, I.N, Sablin, E.P, Xu, R.X, Waitt, G.M, Juzumiene, D, Williams, J.D, Ingraham, H.A, Willson, T.M, Williams, S.P, Montana, V, Madauss, K.P, Moore, J, Bynum, J.M, Lebedeva, L, MacKay, J.A, Suzawa, M, Guy, R.K, Thornton, J.W. | | Deposit date: | 2005-01-28 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1
Cell(Cambridge,Mass.), 120, 2005
|
|
7KM4
 
 | |
4HDS
 
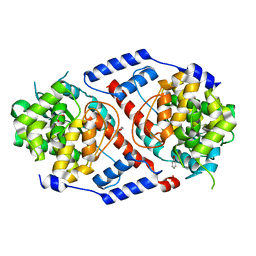 | | Crystal Structure of ArsAB in Complex with Phenol. | | Descriptor: | 1,2-ETHANEDIOL, ArsA, ArsB, ... | | Authors: | Newmister, S.A, Chan, C.H, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Function of the Nicotinate Mononucleotide:phenol/p-cresol Phosphoribosyltransferase (ArsAB) Enzyme from Sporomusa ovata.
Biochemistry, 51, 2012
|
|
1F7H
 
 | |
1F7F
 
 | |
2UV1
 
 | |
2V1C
 
 | |
1F7S
 
 | | CRYSTAL STRUCTURE OF ADF1 FROM ARABIDOPSIS THALIANA | | Descriptor: | ACTIN DEPOLYMERIZING FACTOR (ADF), LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Bowman, G.D, Nodelman, I.M, Lindberg, U, Chua, N.H, Schutt, C.E. | | Deposit date: | 2000-06-27 | | Release date: | 2000-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A comparative structural analysis of the ADF/cofilin family.
Proteins, 41, 2000
|
|
2V4U
 
 | | Human CTP synthetase 2 - glutaminase domain in complex with 5-OXO-L- NORLEUCINE | | Descriptor: | CTP SYNTHASE 2 | | Authors: | Welin, M, Moche, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wisniewska, M, Weigelt, J, Wikstrom, M, Nordlund, P. | | Deposit date: | 2008-09-29 | | Release date: | 2008-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human Ctp Synthetase 2 - Glutaminase Domain in Complex with 5-Oxo-L-Norleucine
To be Published
|
|
7KP2
 
 | | High Resolution Crystal Structure of Putative Pterin Binding Protein (PruR) from Vibrio cholerae O1 biovar El Tor str. N16961 in Complex with Neopterin | | Descriptor: | L-NEOPTERIN, Putative Pterin Binding Protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Pshenychnyi, S, Dubrovska, I, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-10 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | High Resolution Crystal Structure of Putative Pterin Binding Protein (PruR) from Vibrio cholerae O1 biovar El Tor str. N16961 in Complex with Neopterin.
To Be Published
|
|
