3MJ8
 
 | |
2OOM
 
 | | NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA/RNA aptamer | | Descriptor: | RNA 16-mer, TAR RNA element of HIV-1 | | Authors: | Lebars, I, Richard, T, Di Primo, C, Toulme, J.J. | | Deposit date: | 2007-01-26 | | Release date: | 2008-01-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA-modified aptamer
Nucleic Acids Res., 35, 2007
|
|
2OPV
 
 | |
5NFG
 
 | | Structure of recombinant cardosin B from Cynara cardunculus | | Descriptor: | Procardosin-B,Procardosin-B, alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Pereira, P.J.B, Figueiredo, A.C, Manso, J.A, Almeida, C.M, Simoes, I. | | Deposit date: | 2017-03-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.375 Å) | | Cite: | Functional and structural characterization of synthetic cardosin B-derived rennet.
Appl. Microbiol. Biotechnol., 101, 2017
|
|
5NHL
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | (6~{R})-5-(2-methoxyethyl)-6-methyl-2-[5-methyl-2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NHX
 
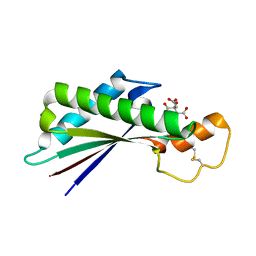 | | Periplasmic domain of Outer Membrane Protein A from Klebsiella pneumoniae | | Descriptor: | CITRIC ACID, Outer membrane protein A | | Authors: | Demange, P, Tranier, S, Nars, G, Iordanov, I, Mourey, L, Saurel, O, Milon, A. | | Deposit date: | 2017-03-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and dynamics of the C-terminal domain of OmpA from Klebsiella pneumonia
To Be Published
|
|
2W4J
 
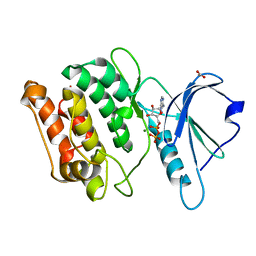 | | X-ray structure of a DAP-Kinase 2-277 | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, DEATH-ASSOCIATED PROTEIN KINASE 1, ... | | Authors: | De Diego, I, Kuper, J, Lehmann, F, Wilmanns, M. | | Deposit date: | 2008-11-27 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-Ray Structure of a Dap-Kinase Calmodulin Complex
To be Published
|
|
2ORL
 
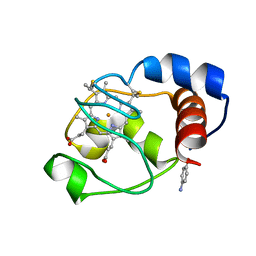 | | Solution structure of the cytochrome c- para-aminophenol adduct | | Descriptor: | 4-AMINOPHENOL, Cytochrome c iso-1, HEME C | | Authors: | Assfalg, M, Bertini, I, Del Conte, R, Giachetti, A, Turano, P. | | Deposit date: | 2007-02-03 | | Release date: | 2007-04-24 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Cytochrome c and organic molecules: solution structure of the p-aminophenol adduct.
Biochemistry, 46, 2007
|
|
3UDJ
 
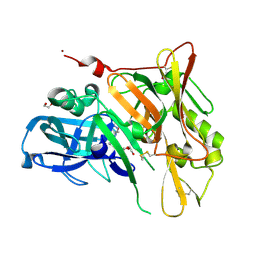 | | Crystal Structure of BACE with Compound 5 | | Descriptor: | 1,2-ETHANEDIOL, Beta-secretase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
3UDR
 
 | | Crystal Structure of BACE with Compound 14 | | Descriptor: | 1,2-ETHANEDIOL, 1-cyanocyclohexyl (3S,5'R)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxylate, Beta-secretase 1, ... | | Authors: | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-18 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
2ONU
 
 | | Plasmodium falciparum ubiquitin conjugating enzyme PF10_0330, putative homologue of human UBE2H | | Descriptor: | TETRAETHYLENE GLYCOL, Ubiquitin-conjugating enzyme, putative | | Authors: | Dong, A, Lew, J, Lin, L, Hassanali, A, Zhao, Y, Senisterra, G, Wasney, G, Vedadi, M, Kozieradzki, I, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Hui, R, Brokx, S.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-24 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Plasmodium falciparum ubiquitin conjugating enzyme PF10_0330, putative homologue of human UBE2H
To be Published
|
|
2OP2
 
 | |
2OWA
 
 | | Crystal structure of putative GTPase activating protein for ADP ribosylation factor from Cryptosporidium parvum (cgd5_1040) | | Descriptor: | Arfgap-like finger domain containing protein, ZINC ION | | Authors: | Dong, A, Lew, J, Zhao, Y, Hassanali, A, Lin, L, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-15 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative GTPase activating protein for ADP ribosylation factor from Cryptosporidium parvum (cgd5_1040)
To be Published
|
|
1CI9
 
 | | DFP-INHIBITED ESTERASE ESTB FROM BURKHOLDERIA GLADIOLI | | Descriptor: | DIISOPROPYL PHOSPHONATE, PROTEIN (CARBOXYLESTERASE) | | Authors: | Wagner, U.G, Petersen, E.I, Schwab, H, Kratky, C. | | Deposit date: | 1999-04-08 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | EstB from Burkholderia gladioli: a novel esterase with a beta-lactamase fold reveals steric factors to discriminate between esterolytic and beta-lactam cleaving activity
Protein Sci., 11, 2002
|
|
5NA7
 
 | | Structure of DPP III from Bacteroides thetaiotaomicron | | Descriptor: | CHLORIDE ION, Putative dipeptidyl-peptidase III, SULFATE ION, ... | | Authors: | Sabljic, I, Luic, M. | | Deposit date: | 2017-02-27 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of dipeptidyl peptidase III from the human gut symbiont Bacteroides thetaiotaomicron.
PLoS ONE, 12, 2017
|
|
5NHI
 
 | |
2WBL
 
 | | Three-dimensional structure of a binary ROP-PRONE complex | | Descriptor: | RAC-LIKE GTP-BINDING PROTEIN ARAC2, RHO OF PLANTS GUANINE NUCLEOTIDE EXCHANGE FACTOR 8 | | Authors: | Thomas, C, Fricke, I, Weyand, M, Berken, A. | | Deposit date: | 2009-03-02 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 3D Structure of a Binary Rop-Prone Complex: The Final Intermediate for a Complete Set of Molecular Snapshots of the Ropgef Reaction.
Biol.Chem., 390, 2009
|
|
2N9H
 
 | | Glucose as a nuclease mimic in DNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*CP*(GL6)P*GP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*CP*TP*GP*CP*TP*AP*G)-3') | | Authors: | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avino, A, Eritja, R, Gonzalez-Ibanez, C, Morales, J, Muro, A, Penalver, P, Fonseca-Guerra, C, Bickelhaupt, M. | | Deposit date: | 2015-11-25 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Glucose-Nucleobase Pseudo Base Pairs: Biomolecular Interactions within DNA.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
1FRD
 
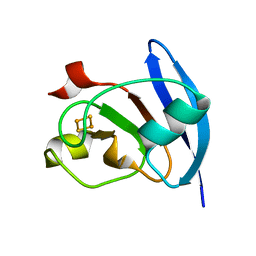 | | MOLECULAR STRUCTURE OF THE OXIDIZED, RECOMBINANT, HETEROCYST (2FE-2S) FERREDOXIN FROM ANABAENA 7120 DETERMINED TO 1.7 ANGSTROMS RESOLUTION | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, HETEROCYST [2FE-2S] FERREDOXIN | | Authors: | Jacobson, B.L, Chae, Y.K, Markley, J.L, Rayment, I, Holden, H.M. | | Deposit date: | 1993-04-14 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular structure of the oxidized, recombinant, heterocyst [2Fe-2S] ferredoxin from Anabaena 7120 determined to 1.7-A resolution.
Biochemistry, 32, 1993
|
|
5NHF
 
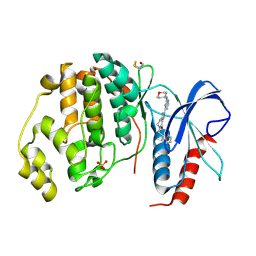 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 2-[2-(oxan-4-ylamino)pyrimidin-4-yl]-5-(phenylmethyl)-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NHJ
 
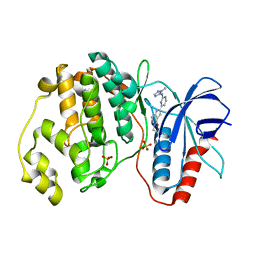 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 5-(2-methoxyethyl)-2-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NHP
 
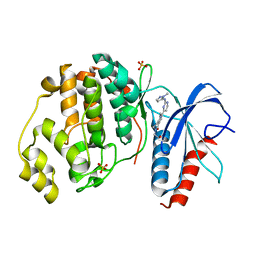 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 5-(2-methoxyethyl)-1-methyl-2-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydropyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
1FAD
 
 | | DEATH DOMAIN OF FAS-ASSOCIATED DEATH DOMAIN PROTEIN, RESIDUES 89-183 | | Descriptor: | PROTEIN (FADD PROTEIN) | | Authors: | Jeong, E.-J, Bang, S, Lee, T.H, Park, Y.-I, Sim, W.-S, Kim, K.-S. | | Deposit date: | 1999-03-23 | | Release date: | 1999-07-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of FADD death domain. Structural basis of death domain interactions of Fas and FADD.
J.Biol.Chem., 274, 1999
|
|
3UMH
 
 | | X-ray structure of the E2 domain of the human amyloid precursor protein (APP) in complex with cadmium | | Descriptor: | ACETATE ION, Amyloid beta A4 protein, CADMIUM ION | | Authors: | Dahms, S.O, Konnig, I, Roeser, D, Guhrs, K.H, Than, M.E. | | Deposit date: | 2011-11-13 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal Binding Dictates Conformation and Function of the Amyloid Precursor Protein (APP) E2 Domain.
J.Mol.Biol., 416, 2012
|
|
5AJR
 
 | | Sterol 14-alpha demethylase (CYP51) from Trypanosoma cruzi in complex with the 1-tetrazole derivative VT-1161 ((R)-2-(2,4-Difluorophenyl)-1, 1-difluoro-3-(1H-tetrazol-1-yl)-1-(5-(4-(2,2,2-trifluoroethoxy)phenyl) pyridin-2-yl)propan-2-ol) | | Descriptor: | (R)-2-(2,4-Difluorophenyl)-1,1-difluoro-3-(1H-tetrazol-1-yl)-1-(5-(4-(2,2,2-trifluoroethoxy)phenyl)pyridin-2-yl)propan-2-ol, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Wawrzak, Z, Hoekstra, W, I Lepesheva, G. | | Deposit date: | 2015-02-26 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Clinical Candidate Vt-1161'S Antiparasitic Effect in Vitro, Activity in a Murine Model of Chagas Disease, and Structural Characterization in Complex with the Target Enzyme Cyp51 from Trypanosoma Cruzi.
Antimicrob.Agents Chemother., 60, 2015
|
|
