1IZ6
 
 | | Crystal Structure of Translation Initiation Factor 5A from Pyrococcus Horikoshii | | Descriptor: | Initiation Factor 5A | | Authors: | Yao, M, Ohsawa, A, Kikukawa, S, Tanaka, I, Kimura, M. | | Deposit date: | 2002-09-25 | | Release date: | 2003-01-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Hyperthermophilic Archaeal Initiation Factor 5A: A Homologue of Eukaryotic Initiation Factor 5A (eIF-5A)
J.BIOCHEM.(TOKYO), 133, 2003
|
|
6JF5
 
 | |
1IEK
 
 | | SOLUTION STRUCTURE OF THE DNA DUPLEX D(CCACCGGAAC).(GTTCCGGTGG) WITH A CHIRAL ALKYL-PHOSPHONATE MOIETY (DIAESTEREOISOMER S) | | Descriptor: | 5'-D(*CP*CP*AP*CP*CP*(OCT)GP*GP*AP*AP*C)-3', 5'-D(*GP*TP*TP*CP*CP*GP*GP*TP*GP*G)-3', N-OCTANE | | Authors: | Soliva, R, Monaco, V, Gomez-Pinto, I, Meeuwenoord, N.J, van der Marel, G.A, van Boom, J.H, Gonzalez, C, Orozco, M. | | Deposit date: | 2001-04-10 | | Release date: | 2001-07-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA duplex with a chiral alkyl phosphonate moiety.
Nucleic Acids Res., 29, 2001
|
|
2BSC
 
 | | E. coli F17a-G lectin domain complex with N-acetylglucosamine, high- resolution structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F17A-G ADHESIN | | Authors: | Buts, L, Wellens, A, Van Molle, I, Wyns, L, Loris, R, Lahmann, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2005-05-20 | | Release date: | 2006-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Impact of Natural Variation in Bacterial F17G Adhesins on Crystallization Behaviour.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4UZS
 
 | | Crystal structure of Bifidobacterium bifidum beta-galactosidase | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Godoy, A.S, Murakami, M.T, Camilo, C.M, Bernardes, A, Polikarpov, I. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of Beta1-6-Galactosidase from Bifidobacterium Bifidum S17: Trimeric Architecture, Molecular Determinants of the Enzymatic Activity and its Inhibition by Alpah-Galactose.
FEBS J., 283, 2016
|
|
1S7H
 
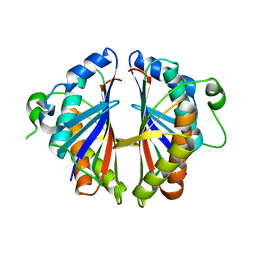 | | Structural Genomics, 2.2A crystal structure of protein YKOF from Bacillus subtilis | | Descriptor: | ykoF | | Authors: | Zhang, R, Lezondra, L, Moy, S, Dementieva, I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-29 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2A crystal structure of protein YKOF from Bacillus subtilis
To be Published
|
|
1S8N
 
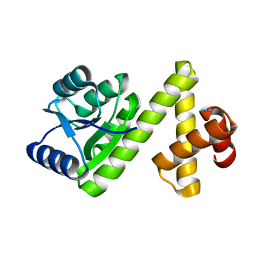 | | Crystal structure of Rv1626 from Mycobacterium tuberculosis | | Descriptor: | AZIDE ION, putative antiterminator | | Authors: | Morth, J.P, Feng, V, Perry, L.J, Svergun, D.I, Tucker, P.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-03 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | The Crystal and Solution Structure of a Putative Transcriptional Antiterminator from Mycobacterium tuberculosis.
Structure, 12, 2004
|
|
2C5Y
 
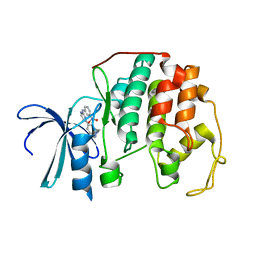 | | DIFFERENTIAL BINDING OF INHIBITORS TO ACTIVE AND INACTIVE CDK2 PROVIDES INSIGHTS FOR DRUG DESIGN | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, HYDROXY(OXO)(3-{[(2Z)-4-[3-(1H-1,2,4-TRIAZOL-1-YLMETHYL)PHENYL]PYRIMIDIN-2(5H)-YLIDENE]AMINO}PHENYL)AMMONIUM | | Authors: | Kontopidis, G, McInnes, C, Pandalaneni, S.R, McNae, I, Gibson, D, Mezna, M, Thomas, M, Wood, G, Wang, S, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2005-11-03 | | Release date: | 2006-03-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Differential Binding of Inhibitors to Active and Inactive Cdk2 Provides Insights for Drug Design.
Chem.Biol., 13, 2006
|
|
5VQJ
 
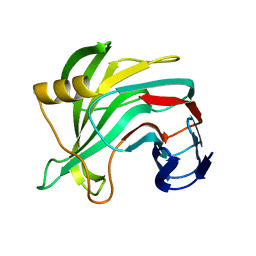 | |
3LDI
 
 | | Crystal structure of aprotinin in complex with sucrose octasulfate: unusual interactions and implication for heparin binding | | Descriptor: | GLYCEROL, MERCURY (II) ION, Pancreatic trypsin inhibitor, ... | | Authors: | Yang, I.S, Kim, T.G, Park, B.S, Kim, K.H. | | Deposit date: | 2010-01-13 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of aprotinin and its complex with sucrose octasulfate reveal multiple modes of interactions with implications for heparin binding
Biochem.Biophys.Res.Commun., 397, 2010
|
|
5VH6
 
 | | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis. | | Descriptor: | CHLORIDE ION, Elongation factor G | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis.
To Be Published
|
|
5VTG
 
 | |
6Q9F
 
 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) H679A in complex with Mn, NOG and Factor X peptide fragment (39mer-4Ser) | | Descriptor: | Aspartyl/asparaginyl beta-hydroxylase, Coagulation factor X, GLYCEROL, ... | | Authors: | Chowdhury, R, Pfeffer, I, Schofield, C.J. | | Deposit date: | 2018-12-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Aspartyl/Asparaginyl beta-hydroxylase (AspH) H679A in complex with Mn, NOG and Factor X peptide fragment (39mer-4Ser)
To Be Published
|
|
6Q9I
 
 | |
2BX8
 
 | | Human serum albumin complexed with azapropazone | | Descriptor: | AZAPROPAZONE, SERUM ALBUMIN | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | Deposit date: | 2005-07-25 | | Release date: | 2005-09-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
3LII
 
 | | Recombinant human acetylcholinesterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, SULFATE ION | | Authors: | Dvir, H, Rosenberry, T, Harel, M, Silman, I, Sussman, J. | | Deposit date: | 2010-01-25 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Acetylcholinesterase: From 3D structure to function.
Chem.Biol.Interact, 187, 2010
|
|
1IEY
 
 | | SOLUTION STRUCTURE OF THE DNA DUPLEX D(CCACCGGAAC).(GTTCCGGTGG) WITH A CHIRAL ALKYL-PHOSPHONATE MOIETY (DIAESTEREOISOMER R) | | Descriptor: | 5'-D(*GP*TP*TP*CP*CP*GP*GP*TP*GP*G)-3', 5'-D(P*CP*CP*AP*CP*CP*(OCT)GP*GP*AP*AP*C)-3', N-OCTANE | | Authors: | Soliva, R, Monaco, V, Gomez-Pinto, I, Meeuwenoord, N.J, van der Marel, G.A, van Boom, J.H, Gonzalez, C, Orozco, M. | | Deposit date: | 2001-04-11 | | Release date: | 2001-07-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA duplex with a chiral alkyl phosphonate moiety.
Nucleic Acids Res., 29, 2001
|
|
1IF7
 
 | | Carbonic Anhydrase II Complexed With (R)-N-(3-Indol-1-yl-2-methyl-propyl)-4-sulfamoyl-benzamide | | Descriptor: | (R)-N-(3-INDOL-1-YL-2-METHYL-PROPYL)-4-SULFAMOYL-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Grzybowski, B.A, Ishchenko, A.V, Kim, C.-Y, Topalov, G, Chapman, R, Christianson, D.W, Whitesides, G.M, Shakhnovich, E.I. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Combinatorial computational method gives new picomolar ligands for a known enzyme.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5VLQ
 
 | | Structure of the TTLL3 Glycylase | | Descriptor: | LOC100158544 protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION | | Authors: | Garnham, C.P, Yu, I, Li, Y, Roll-Mecak, A. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Crystal structure of tubulin tyrosine ligase-like 3 reveals essential architectural elements unique to tubulin monoglycylases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1IFQ
 
 | | Sec22b N-terminal domain | | Descriptor: | GLYCEROL, vesicle trafficking protein Sec22b | | Authors: | Gonzalez Jr, L.C, Weis, W.I, Scheller, R.H. | | Deposit date: | 2001-04-13 | | Release date: | 2001-05-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel snare N-terminal domain revealed by the crystal structure of Sec22b.
J.Biol.Chem., 276, 2001
|
|
4U8M
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Y317A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
3LOF
 
 | | C-terminal domain of human heat shock 70kDa protein 1B. | | Descriptor: | Heat shock 70 kDa protein 1 | | Authors: | Osipiuk, J, Gu, M, Mihelic, M, Orton, K, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-03 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of C-terminal domain of human heat shock 70kDa protein 1B.
To be Published
|
|
1J3A
 
 | | Crystal structure of ribosomal protein L13 from Pyrococcus horikoshii | | Descriptor: | 50S ribosomal protein L13P | | Authors: | Nakashima, T, Tanaka, M, Kazama, T, Kawamura, S, Kimura, M, Yao, M, Tanaka, I. | | Deposit date: | 2003-01-21 | | Release date: | 2003-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of ribosomal protein L13 from hyperthermophilic archaeon Pyrococcus horikoshii
To be Published
|
|
1IHC
 
 | | X-ray Structure of Gephyrin N-terminal Domain | | Descriptor: | Gephyrin | | Authors: | Sola, M, Kneussel, M, Heck, I.S, Betz, H, Weissenhorn, W. | | Deposit date: | 2001-04-21 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystal structure of the trimeric N-terminal domain of gephyrin.
J.Biol.Chem., 276, 2001
|
|
2BXA
 
 | | Human serum albumin complexed with 3-carboxy-4-methyl-5-propyl-2- furanpropanoic acid (CMPF) | | Descriptor: | 3-CARBOXY-4-METHYL-5-PROPYL-2-FURANPROPIONIC, SERUM ALBUMIN | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Otagiri, M, Curry, S. | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
