2ADV
 
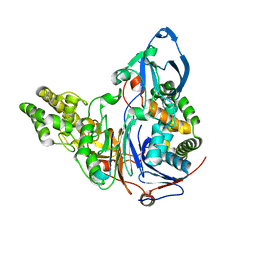 | | Crystal Structures Of Glutaryl 7-Aminocephalosporanic Acid Acylase: mutational study of activation mechanism | | Descriptor: | Glutaryl 7- Aminocephalosporanic Acid Acylase | | Authors: | Kim, J.K, Yang, I.S, Shin, H.J, Cho, K.J, Ryu, E.K, Kim, S.H, Park, S.S, Kim, K.H. | | Deposit date: | 2005-07-21 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Insight into autoproteolytic activation from the structure of cephalosporin acylase: a protein with two proteolytic chemistries.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2AFE
 
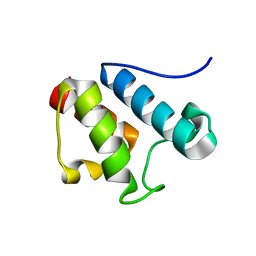 | | Solution Structure of Asl1650, an Acyl Carrier Protein from Anabaena sp. PCC 7120 with a Variant Phosphopantetheinylation-Site Sequence | | Descriptor: | protein Asl1650 | | Authors: | Johnson, M.A, Peti, W, Herrmann, T, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2005-07-25 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Asl1650, an acyl carrier protein from Anabaena sp. PCC 7120 with a variant phosphopantetheinylation-site sequence
Protein Sci., 15, 2006
|
|
2AEM
 
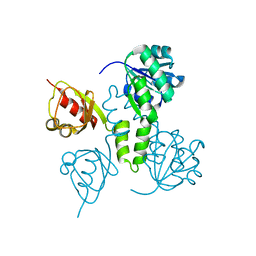 | | Crystal Structures of the MthK RCK Domain | | Descriptor: | Calcium-gated potassium channel mthK | | Authors: | Dong, J, Shi, N, Berke, I, Chen, L, Jiang, Y. | | Deposit date: | 2005-07-22 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the MthK RCK Domain and the Effect of Ca2+ on Gating Ring Stability
J.Biol.Chem., 280, 2005
|
|
3KY2
 
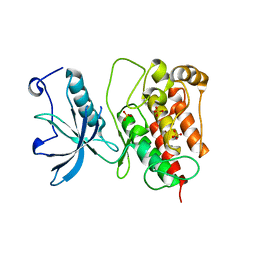 | | Crystal structure of Fibroblast Growth Factor Receptor 1 kinase domain | | Descriptor: | Basic fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Bae, J.H, Boggon, T.J, Tome, F, Mandiyan, V, Lax, I, Schlessinger, J. | | Deposit date: | 2009-12-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Asymmetric receptor contact is required for tyrosine autophosphorylation of fibroblast growth factor receptor in living cells.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KYI
 
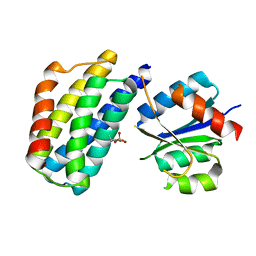 | | Crystal structure of the phosphorylated P1 domain of CheA3 in complex with CheY6 from R. sphaeroides | | Descriptor: | CheY6 protein, Putative histidine protein kinase | | Authors: | Bell, C.H, Porter, S.L, Armitage, J.P, Stuart, D.I. | | Deposit date: | 2009-12-06 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Using structural information to change the phosphotransfer specificity of a two-component chemotaxis signalling complex
Plos Biol., 8, 2010
|
|
1OY9
 
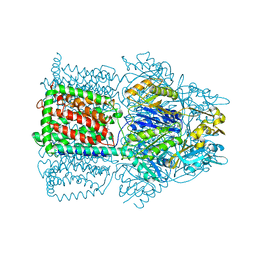 | | Structural Basis of Multiple Drug Binding Capacity of the AcrB Multidrug Efflux Pump | | Descriptor: | Acriflavine resistance protein B, ETHIDIUM | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
1J5D
 
 | | SOLUTION STRUCTURE OF OXIDIZED PARAMAGNETIC CU(II) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803-MINIMIZED AVERAGE STRUCTURE | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Shumilin, S, Vila, A.J. | | Deposit date: | 2002-04-02 | | Release date: | 2002-04-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The first solution structure of a paramagnetic copper(II) protein: the case of oxidized plastocyanin from the cyanobacterium Synechocystis PCC6803.
J.Am.Chem.Soc., 123, 2001
|
|
1ZX6
 
 | | High-resolution crystal structure of yeast Pin3 SH3 domain | | Descriptor: | Ypr154wp | | Authors: | Kursula, P, Kursula, I, Lehmann, F, Zou, P, Song, Y.H, Wilmanns, M. | | Deposit date: | 2005-06-07 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural genomics of yeast SH3 domains
To be Published
|
|
241D
 
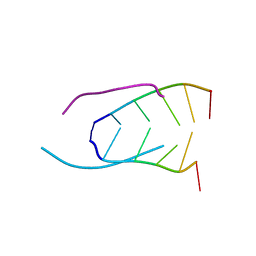 | | EXTENSION OF THE FOUR-STRANDED INTERCALATED CYTOSINE MOTIF BY ADENINE.ADENINE BASE PAIRING IN THE CRYSTAL STRUCTURE OF D(CCCAAT) | | Descriptor: | DNA (5'-D(*CP*CP*CP*AP*AP*T)-3') | | Authors: | Berger, I, Kang, C, Fredian, A, Ratliff, R, Moyzis, R, Rich, A. | | Deposit date: | 1995-12-07 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Extension of the four-stranded intercalated cytosine motif by adenine.adenine base pairing in the crystal structure of d(CCCAAT).
Nat.Struct.Biol., 2, 1995
|
|
2A08
 
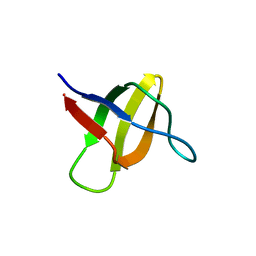 | | Structure of the yeast YHH6 SH3 domain | | Descriptor: | Hypothetical 41.8 kDa protein in SPO13-ARG4 intergenic region | | Authors: | Kursula, P, Kursula, I, Song, Y.H, Lehmann, F, Zou, P, Wilmanns, M. | | Deposit date: | 2005-06-16 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | 3-D proteome of yeast SH3 domains
To be Published
|
|
1JJF
 
 | | STRUCTURAL BASIS FOR THE SUBSTRATE SPECIFICITY OF THE FERULOYL ESTERASE DOMAIN OF THE CELLULOSOMAL XYLANASE Z OF CLOSTRIDIUM THERMOCELLUM | | Descriptor: | ENDO-1,4-BETA-XYLANASE Z, PLATINUM (II) ION | | Authors: | Schubot, F.D, Kataeva, I.A, Blum, D.L, Shah, A.K, Ljungdahl, L.G, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2001-07-05 | | Release date: | 2001-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the substrate specificity of the feruloyl esterase domain of the cellulosomal xylanase Z from Clostridium thermocellum.
Biochemistry, 40, 2001
|
|
1QRW
 
 | | CRYSTAL STRUCTURE OF AN ALPHA-LYTIC PROTEASE MUTANT WITH ACCELERATED FOLDING KINETICS, R102H/G134S, PH 8 | | Descriptor: | ALHPA-LYTIC PROTEASE, GLYCEROL, SULFATE ION | | Authors: | Derman, A.I, Mau, T, Agard, D.A. | | Deposit date: | 1999-06-16 | | Release date: | 1999-06-18 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Genetic Screen That Targets Specifically the Folding Transition State of Alpha-Lytic Protease
To be Published, 1999
|
|
1K1U
 
 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, W Harrison, R, Weber, I.T. | | Deposit date: | 2001-09-25 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
1QYK
 
 | | GCATGCT + Barium | | Descriptor: | 5'-D(*GP*CP*AP*TP*GP*CP*T)-3', BARIUM ION | | Authors: | Cardin, C.J, Gan, Y, Thorpe, J.H, Teixeira, S.C.M, Gale, B.C, Moraes, M.I.A. | | Deposit date: | 2003-09-11 | | Release date: | 2003-10-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Metal Ion Distribution and Stabilization of the DNA Quadruplex Structure Formed by d(GCATGCT)
To be published
|
|
1Q6O
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-gulonaet 6-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-GULURONIC ACID 6-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-08-13 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.202 Å) | | Cite: | Structural Evidence for a 1,2-Enediolate Intermediate in the Reaction Catalyzed by 3-Keto-l-Gulonate 6-Phosphate Decarboxylase, a Member of the Orotidine 5'-Monophosphate Decarboxylase Suprafamily
Biochemistry, 42, 2003
|
|
3KY7
 
 | | 2.35 Angstrom resolution crystal structure of a putative tRNA (guanine-7-)-methyltransferase (trmD) from Staphylococcus aureus subsp. aureus MRSA252 | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Dubrovska, I, Shuvalova, L, See, R, Zoraghi, R, Reiner, N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-04 | | Release date: | 2009-12-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution crystal structure of a putative tRNA (guanine-7-)-methyltransferase (trmD) from Staphylococcus aureus subsp. aureus MRSA252
To be Published
|
|
1JHR
 
 | | Three-dimensional Structure of CobT in Complex with Reaction Products of 2-hydroxypurine and NaMN | | Descriptor: | N7-(5'-PHOSPHO-ALPHA-RIBOSYL)-2-HYDROXYPURINE, NICOTINIC ACID, Nicotinate Mononucleotide:5,6-Dimethylbenzimidazole Phosphoribosyltransferase | | Authors: | Cheong, C.G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica.
J.Biol.Chem., 276, 2001
|
|
1YXK
 
 | | Crystal structure of human lectin-like oxidized low-density lipoprotein receptor 1 (LOX-1) disulfide-linked dimer | | Descriptor: | oxidised low density lipoprotein (lectin-like) receptor 1 | | Authors: | Ohki, I, Ishigaki, T, Oyama, T, Matsunaga, S, Xie, Q, Ohnishi-Kameyama, M, Murata, T, Tsuchiya, D, Machida, S, Morikawa, K, Tate, S. | | Deposit date: | 2005-02-22 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human lectin-like, oxidized low-density lipoprotein receptor 1 ligand binding domain and its ligand recognition mode to OxLDL.
Structure, 13, 2005
|
|
1QCI
 
 | | LOW TEMPERATURE STRUCTURE OF POKEWEED ANTIVIRAL PROTEIN COMPLEXED WITH ADENINE | | Descriptor: | ADENINE, POKEWEED ANTIVIRAL PROTEIN | | Authors: | Kurinov, I.V, Myers, D.E, Irvin, J.D, Uckun, F.M. | | Deposit date: | 1999-05-05 | | Release date: | 1999-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic analysis of the structural basis for the interactions of pokeweed antiviral protein with its active site inhibitor and ribosomal RNA substrate analogs.
Protein Sci., 8, 1999
|
|
4JY5
 
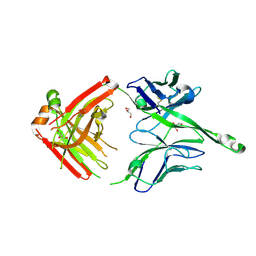 | | Crystal structure of human Fab PGT122, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | GLYCEROL, PGT122 heavy chain, PGT122 light chain | | Authors: | Julien, J.-P, Diwanji, D.C, Burton, D.R, Wilson, I.A. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans.
Plos Pathog., 9, 2013
|
|
1JQZ
 
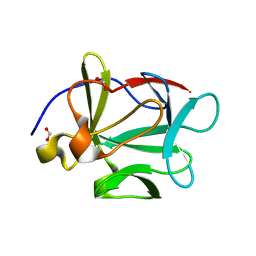 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Terminal His Tag. | | Descriptor: | FORMIC ACID, acidic fibroblast growth factor | | Authors: | Brych, S.R, Blaber, S.I, Logan, T.M, Blaber, M. | | Deposit date: | 2001-08-09 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core
region of a beta-trefoil.
Protein Sci., 10, 2001
|
|
1JT9
 
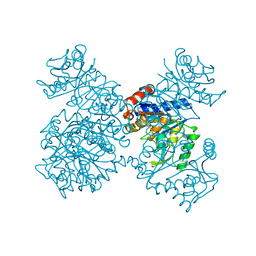 | | Structure of the mutant F174A T form of the Glucosamine-6-Phosphate deaminase from E.coli | | Descriptor: | Glucosamine-6-Phosphate deaminase | | Authors: | Bustos-Jaimes, I, Sosa-Peinado, A, Rudino-Pinera, E, Horjales, E, Calcagno, M.L. | | Deposit date: | 2001-08-20 | | Release date: | 2002-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | On the role of the conformational flexibility of the active-site lid on the allosteric kinetics of glucosamine-6-phosphate deaminase.
J.Mol.Biol., 319, 2002
|
|
1Q7G
 
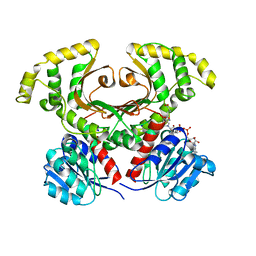 | | Homoserine Dehydrogenase in complex with suicide inhibitor complex NAD-5-hydroxy-4-Oxonorvaline | | Descriptor: | Homoserine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE-5-HYDROXY-4-OXONORVALINE, SODIUM ION | | Authors: | Jacques, S.L, Mirza, I.A, Ejim, L, Koteva, K, Hughes, D.W, Green, K, Kinach, R, Honek, J.F, Lai, H.K, Berghuis, A.M, Wright, G.D. | | Deposit date: | 2003-08-18 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Enzyme assisted suicide: Molecular basis for the antifungal activity of 5-hydroxy-4-oxonorvaline by potent inhibition of homoserine dehydrogenase
Chem.Biol., 10, 2003
|
|
3LNB
 
 | | Crystal Structure Analysis of Arylamine N-acetyltransferase C from Bacillus anthracis | | Descriptor: | COENZYME A, FORMIC ACID, N-acetyltransferase family protein | | Authors: | Li de la Sierra-Gallay, I, Pluvinage, B, Rodrigues-Lima, F. | | Deposit date: | 2010-02-02 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Bacillus anthracis arylamine N-acetyltransferase ((BACAN)NAT1) that inactivates sulfamethoxazole, reveals unusual structural features compared with the other NAT isoenzymes.
Febs Lett., 585, 2011
|
|
1Q6R
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-xylulose 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-XYLULOSE 5-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-08-13 | | Release date: | 2003-10-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Evidence for a 1,2-Enediolate Intermediate in the Reaction Catalyzed by 3-Keto-l-Gulonate 6-Phosphate Decarboxylase, a Member of the Orotidine 5'-Monophosphate Decarboxylase Suprafamily
Biochemistry, 42, 2003
|
|
