6AW9
 
 | | 2.55A resolution structure of SAH bound catechol O-methyltransferase (COMT) L136M from Nannospalax galili | | Descriptor: | CALCIUM ION, Catechol O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
4DST
 
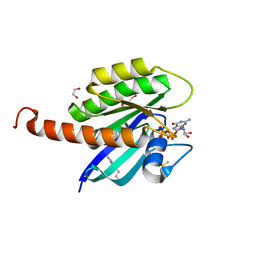 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | 1,2-ETHANEDIOL, 2-(4,6-dichloro-2-methyl-1H-indol-3-yl)ethanamine, ACETATE ION, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5NHI
 
 | |
5NA7
 
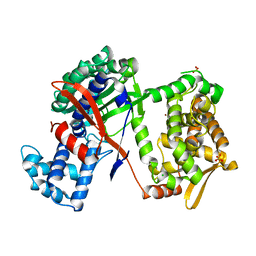 | | Structure of DPP III from Bacteroides thetaiotaomicron | | Descriptor: | CHLORIDE ION, Putative dipeptidyl-peptidase III, SULFATE ION, ... | | Authors: | Sabljic, I, Luic, M. | | Deposit date: | 2017-02-27 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of dipeptidyl peptidase III from the human gut symbiont Bacteroides thetaiotaomicron.
PLoS ONE, 12, 2017
|
|
6QUZ
 
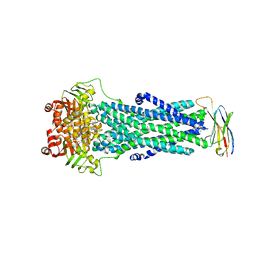 | | Structure of ATPgS-bound outward-facing TM287/288 in complex with sybody Sb_TM35 | | Descriptor: | ABC transporter, ATP-binding protein, MAGNESIUM ION, ... | | Authors: | Hutter, C.A.J, Huerlimann, L.M, Zimmermann, I, Egloff, P, Seeger, M.A. | | Deposit date: | 2019-03-01 | | Release date: | 2019-05-29 | | Last modified: | 2019-06-05 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | The extracellular gate shapes the energy profile of an ABC exporter.
Nat Commun, 10, 2019
|
|
8ATB
 
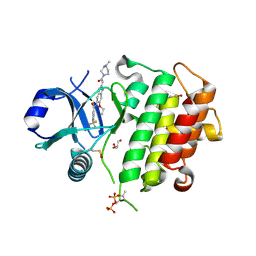 | | Discovery of IRAK4 Inhibitor 16 | | Descriptor: | GLYCEROL, Interleukin-1 receptor-associated kinase 4, ~{N}-[6-ethoxy-2-[2-(4-methylpiperazin-1-yl)-2-oxidanylidene-ethyl]indazol-5-yl]-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wenger, A.M, Guimond, N, Thaler, T, Platzek, J, Eberspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-08-22 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
2X6C
 
 | | Potassium Channel from Magnetospirillum Magnetotacticum | | Descriptor: | ATP-SENSITIVE INWARD RECTIFIER POTASSIUM CHANNEL 10, CHLORIDE ION, PHOSPHOCHOLINE, ... | | Authors: | Clarke, O.B, Caputo, A.T, Hill, A.P, Vandenberg, J.I, Smith, B.J, Gulbis, J.M. | | Deposit date: | 2010-02-15 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels.
Cell(Cambridge,Mass.), 141, 2010
|
|
4DPB
 
 | | The 1.00 Angstrom crystal structure of oxidized (CuII) poplar plastocyanin A at pH 8.0 | | Descriptor: | COPPER (II) ION, Plastocyanin A, chloroplastic | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
6QNR
 
 | | 70S ribosome elongation complex (EC) with experimentally assigned potassium ions | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2019-02-11 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction.
Nat Commun, 10, 2019
|
|
1L7M
 
 | | HIGH RESOLUTION LIGANDED STRUCTURE OF PHOSPHOSERINE PHOSPHATASE (PI COMPLEX) | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Phosphoserine Phosphatase | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-15 | | Release date: | 2002-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
6F5P
 
 | |
6P5B
 
 | | Crystal Structure of MavC in Complex with Ub-UbE2N | | Descriptor: | MavC, Ubiquitin, Ubiquitin-conjugating enzyme E2 N | | Authors: | Puvar, K, Iyer, S, Negron Teron, K.I, Das, C. | | Deposit date: | 2019-05-30 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Legionella effector MavC targets the Ube2N~Ub conjugate for noncanonical ubiquitination.
Nat Commun, 11, 2020
|
|
5NHF
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 2-[2-(oxan-4-ylamino)pyrimidin-4-yl]-5-(phenylmethyl)-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
7MWX
 
 | | Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2 with tamarin CD81 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy Chain, ... | | Authors: | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | Deposit date: | 2021-05-17 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
5NHJ
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 5-(2-methoxyethyl)-2-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
7MWS
 
 | | Crystal structure of tamarin CD81 large extracellular loop | | Descriptor: | CD81 protein, GLYCEROL, TETRAETHYLENE GLYCOL | | Authors: | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | Deposit date: | 2021-05-17 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
1P2O
 
 | | Structural consequences of accommodation of four non-cognate amino-acid residues in the S1 pocket of bovine trypsin and chymotrypsin | | Descriptor: | Chymotrypsinogen A, Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Helland, R, Czapinska, H, Leiros, I, Olufsen, M, Otlewski, J, Smalaas, A.O. | | Deposit date: | 2003-04-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural consequences of accommodation of four non-cognate amino acid residues in the S1 pocket of bovine trypsin and chymotrypsin.
J.Mol.Biol., 333, 2003
|
|
5NHP
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 5-(2-methoxyethyl)-1-methyl-2-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydropyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
1P8J
 
 | | CRYSTAL STRUCTURE OF THE PROPROTEIN CONVERTASE FURIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DECANOYL-ARG-VAL-LYS-ARG-CHLOROMETHYLKETONE INHIBITOR, ... | | Authors: | Henrich, S, Cameron, A, Bourenkov, G.P, Kiefersauer, R, Huber, R, Lindberg, I, Bode, W, Than, M.E. | | Deposit date: | 2003-05-07 | | Release date: | 2003-07-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of the Proprotein Processing Proteinase Furin Explains its Stringent Specificity
Nat.Struct.Biol., 10, 2003
|
|
3CF1
 
 | | Structure of P97/vcp in complex with ADP/ADP.alfx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Transitional endoplasmic reticulum ATPase | | Authors: | Davies, J.M, Delabarre, B, Brunger, A.T, Weis, W.I. | | Deposit date: | 2008-03-01 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change.
Structure, 16, 2008
|
|
6PBW
 
 | | Crystal structure of Fab667 complex | | Descriptor: | Fab667 heavy chain, Fab667 light chain, GLYCEROL, ... | | Authors: | Oyen, D, Wilson, I.A. | | Deposit date: | 2019-06-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.058 Å) | | Cite: | Structure and mechanism of monoclonal antibody binding to the junctional epitope of Plasmodium falciparum circumsporozoite protein.
Plos Pathog., 16, 2020
|
|
6BCZ
 
 | | Crystal structure of human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl (2-{[(2R)-3-oxo-2-[(propan-2-yl)amino]-3-{[(pyridin-3-yl)methyl]amino}propyl]sulfanyl}ethyl)carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Inhibition of Human CYP3A4 by Rationally Designed Ritonavir-Like Compounds: Impact and Interplay of the Side Group Functionalities.
Mol Pharm., 15, 2018
|
|
6BD6
 
 | | Crystal structure of human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl (2-{[(2S)-3-oxo-2-(phenylamino)-3-{[(pyridin-3-yl)methyl]amino}propyl]sulfanyl}ethyl)carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2017-10-21 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition of Human CYP3A4 by Rationally Designed Ritonavir-Like Compounds: Impact and Interplay of the Side Group Functionalities.
Mol Pharm., 15, 2018
|
|
6B9P
 
 | | Structure of GH 38 Jack Bean alpha-mannosidase in complex with a 36-valent iminosugar cluster inhibitor | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{9-[4-(methoxymethyl)-1H-1,2,3-triazol-1-yl]nonyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-mannosidase from Canavalia ensiformis (jack bean), ... | | Authors: | Howard, E, Cousido-Siah, A, Lepage, M, Bodlenner, A, Mitschler, A, Meli, A, De Riccardis, F, Izzo, I, Podjarny, A, Compain, P. | | Deposit date: | 2017-10-11 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Structural Basis of Outstanding Multivalent Effects in Jack Bean alpha-Mannosidase Inhibition.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6BDI
 
 | | Crystal structure of human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-(1H-indol-3-yl)-3-{[(2R)-3-oxo-2-[(propan-2-yl)amino]-3-{[(pyridin-3-yl)methyl]amino}propyl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2017-10-23 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Interaction of the rationally designed ritonavir-like inhibitors with human cytochrome P450 3A4: Impact of the side group interplay
Mol. Pharm., 2017
|
|
