4U4R
 
 | | Crystal structure of Lactimidomycin bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R,5S,6E)-2-hydroxy-5-methyl-7-[(2R,3S,4E,6Z,10E)-3-methyl-12-oxooxacyclododeca-4,6,10-trien-2-yl]-4-oxooct-6-en-1-yl}piperidine-2,6-dione, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
1EPF
 
 | | CRYSTAL STRUCTURE OF THE TWO N-TERMINAL IMMUNOGLOBULIN DOMAINS OF THE NEURAL CELL ADHESION MOLECULE (NCAM) | | Descriptor: | CALCIUM ION, PROTEIN (NEURAL CELL ADHESION MOLECULE) | | Authors: | Kasper, C, Rasmussen, H, Kastrup, J.S, Ikemizu, S, Jones, E.Y, Berezin, V, Bock, E, Larsen, I.K. | | Deposit date: | 2000-03-29 | | Release date: | 2000-10-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of cell-cell adhesion by NCAM.
Nat.Struct.Biol., 7, 2000
|
|
4UCF
 
 | | Crystal structure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, N-PROPANOL, ... | | Authors: | Godoy, A.S, Murakami, M.T, Camilo, C.M, Bernardes, A, Polikarpov, I. | | Deposit date: | 2014-12-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Beta1-6-Galactosidase from Bifidobacterium Bifidum S17: Trimeric Architecture, Molecular Determinants of the Enzymatic Activity and its Inhibition by Alpha-Galactose.
FEBS J., 283, 2016
|
|
4F8Z
 
 | | Carboxypeptidase T with Boc-Leu | | Descriptor: | CALCIUM ION, Carboxypeptidase T, GLYCEROL, ... | | Authors: | Timofeev, V.I, Kuznetsov, S.A, Akparov, V.K, Kuranova, I.P. | | Deposit date: | 2012-05-18 | | Release date: | 2013-05-22 | | Last modified: | 2015-09-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Three-dimensional structure of carboxypeptidase T from Thermoactinomyces vulgaris in complex with N-BOC-L-leucine.
Biochemistry Mosc., 78, 2013
|
|
6I8F
 
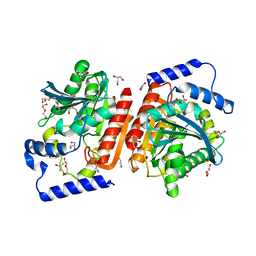 | |
1G5T
 
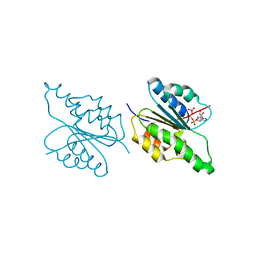 | | THE THREE-DIMENSIONAL STRUCTURE OF ATP:CORRINOID ADENOSYLTRANSFERASE FROM SALMONELLA TYPHIMURIUM. APO-ATP FORM | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COB(I)ALAMIN ADENOSYLTRANSFERASE, MAGNESIUM ION | | Authors: | Rayment, I, Escalante-Semerena, J.C, Bauer, C.B. | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of ATP:corrinoid adenosyltransferase from Salmonella typhimurium in its free state, complexed with MgATP, or complexed with hydroxycobalamin and MgATP.
Biochemistry, 40, 2001
|
|
8C24
 
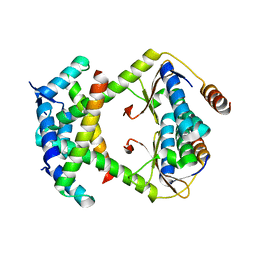 | |
2F8G
 
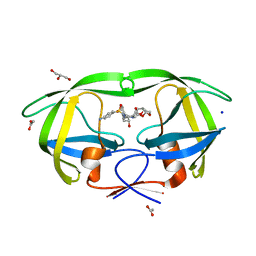 | | HIV-1 protease mutant I50V complexed with inhibitor TMC114 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2005-12-02 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Effectiveness of Nonpeptide Clinical Inhibitor TMC-114 on HIV-1 Protease with Highly Drug Resistant Mutations D30N, I50V, and L90M.
J.Med.Chem., 49, 2006
|
|
3RG1
 
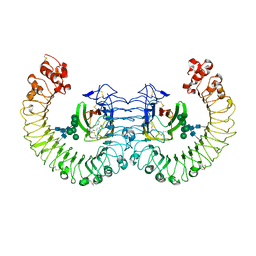 | | Crystal structure of the RP105/MD-1 complex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, CD180 molecule, LY86 protein, ... | | Authors: | Yoon, S.I, Hong, M, Wilson, I.A. | | Deposit date: | 2011-04-07 | | Release date: | 2011-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | An unusual dimeric structure and assembly for TLR4 regulator RP105-MD-1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7NXJ
 
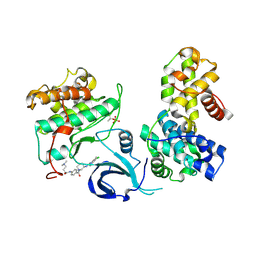 | | Crystal structure of human Cdk13/Cyclin K in complex with the inhibitor THZ531 | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 13, N-[4-[(3R)-3-[[5-chloranyl-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]piperidin-1-yl]carbonylphenyl]-4-(dimethylamino)butanamide | | Authors: | Anand, K, Greifenberg, A.K, Kaltheuner, I.H, Geyer, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-activity relationship study of THZ531 derivatives enables the discovery of BSJ-01-175 as a dual CDK12/13 covalent inhibitor with efficacy in Ewing sarcoma.
Eur.J.Med.Chem., 221, 2021
|
|
3R5R
 
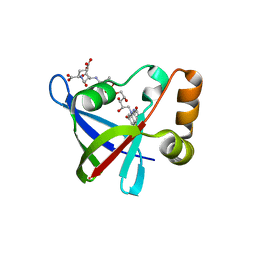 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | Descriptor: | COENZYME F420, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
7NXK
 
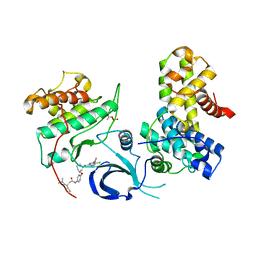 | | Crystal structure of human Cdk12/Cyclin K in complex with the inhibitor BSJ-01-175 | | Descriptor: | (E)-N-[4-[(1R,3R)-3-[[5-chloranyl-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]cyclohexyl]oxyphenyl]-4-(dimethylamino)but-2-enamide, Cyclin-K, Cyclin-dependent kinase 12 | | Authors: | Anand, K, Dust, S, Kaltheuner, I.H, Geyer, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-activity relationship study of THZ531 derivatives enables the discovery of BSJ-01-175 as a dual CDK12/13 covalent inhibitor with efficacy in Ewing sarcoma.
Eur.J.Med.Chem., 221, 2021
|
|
1WN6
 
 | | Crystal Structure of Blasticidin S Deaminase (BSD) Complexed with Tetrahedral Intermediate of Blasticidin S | | Descriptor: | 6-(4-AMINO-4-HYDROXY-2-OXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-3-[3-AMINO-5-(N-METHYL-GUANIDINO)-PENT ANOYLAMINO]-3,6-DIHYDRO-2H-PYRAN-2-CARBOXYLIC ACID, ARSENIC, Blasticidin-S deaminase, ... | | Authors: | Kumasaka, T, Yamamoto, M, Furuichi, M, Nakasako, M, Kimura, M, Yamaguchi, I, Ueki, T. | | Deposit date: | 2004-07-27 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc
J.Biol.Chem., 282, 2007
|
|
4U3M
 
 | | Crystal structure of Anisomycin bound to the yeast 80S ribosome | | Descriptor: | 18S rRNA, 25s rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
6FVF
 
 | | The Structure of CK2alpha with CCh503 bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, [1-[2-(phenylsulfonylamino)ethyl]piperidin-4-yl]methyl 5-fluoranyl-2-methoxy-1~{H}-indole-3-carboxylate | | Authors: | Brear, P, Prudent, R, Laudet, B, Filhol, O, Cochet, C, Sautel, C, Moucadel, V, Bestgen, B, Engel, M, Ettaoussi, M, Lomberget, T, Le Borgne, M, Kufareva, I, Abagyan, R, Hyvonen, M. | | Deposit date: | 2018-03-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of holoenzyme-disrupting chemicals as substrate-selective CK2 inhibitors.
Sci Rep, 9, 2019
|
|
1EKQ
 
 | |
6HCS
 
 | |
7NQZ
 
 | | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with Z1827898537 | | Descriptor: | (3S)-N-benzylpyrrolidin-3-amine, AMP PHOSPHORAMIDATE, CHLORIDE ION, ... | | Authors: | Mohamad, N, O'Donoghue, A, Kantsadi, A.L, Vakonakis, I. | | Deposit date: | 2021-03-02 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | Structures of P. falciparum Hsp70-x nucleotide binding domain with small molecule ligands
To Be Published
|
|
7NQR
 
 | | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with Z287256168 | | Descriptor: | (S)-N-(1-cyclopropylethyl)-6-methylpicolinamide, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, AMP PHOSPHORAMIDATE, ... | | Authors: | Mohamad, N, O'Donoghue, A, Kantsadi, A.L, Vakonakis, I. | | Deposit date: | 2021-03-02 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structures of P. falciparum Hsp70-x nucleotide binding domain with small molecule ligands
To Be Published
|
|
7NQU
 
 | | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with Z396380540 | | Descriptor: | AMP PHOSPHORAMIDATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Mohamad, N, O'Donoghue, A, Kantsadi, A.L, Vakonakis, I. | | Deposit date: | 2021-03-02 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structures of P. falciparum Hsp70-x nucleotide binding domain with small molecule ligands
To Be Published
|
|
7NQS
 
 | | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with Z1203107138 | | Descriptor: | 1-ethyl-N-[(4-fluorophenyl)methyl]-1H-pyrazole-4-carboxamide, AMP PHOSPHORAMIDATE, GLYCEROL, ... | | Authors: | Mohamad, N, O'Donoghue, A, Kantsadi, A.L, Vakonakis, I. | | Deposit date: | 2021-03-02 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structures of P. falciparum Hsp70-x nucleotide binding domain with small molecule ligands
To Be Published
|
|
8F3C
 
 | | Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | Descriptor: | DNA (38-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2022-11-09 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6H9J
 
 | | Factor Inhibiting HIF (FIH) in complex with zinc, NOG and TRPV3 (229-255) | | Descriptor: | GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, N-OXALYLGLYCINE, ... | | Authors: | Leissing, T.M, Clifton, I.J, Saward, B.G, Lu, X, Hopkinson, R.J, Schofield, C.J. | | Deposit date: | 2018-08-04 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Factor Inhibiting HIF (FIH) in complex with zinc, NOG and TRPV3 (229-255)
To Be Published
|
|
4U55
 
 | | Crystal structure of Cryptopleurine bound to the yeast 80S ribosome | | Descriptor: | (14aR)-2,3,6-trimethoxy-11,12,13,14,14a,15-hexahydro-9H-dibenzo[f,h]pyrido[1,2-b]isoquinoline, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U52
 
 | | Crystal structure of Nagilactone C bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
