6YWQ
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148H variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
6YWG
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148N variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
6YXC
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148R variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SODIUM ION | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
8DAO
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV44-79 | | Descriptor: | COV44-79 heavy chain constant domain, COV44-79 heavy chain variable domain, COV44-79 light chain constant domain, ... | | Authors: | Lin, T.H, Lee, C.C.D, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
6F6Q
 
 | |
7UT1
 
 | |
7PL9
 
 | |
7PJM
 
 | | Crystal Structure of Ivosidenib-resistant IDH1 variant R132C S280F in complex with NADPH and Ca2+/2-Oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Reinbold, R, Rabe, P, Abboud, M.I, Schofield, C.J. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Resistance to the isocitrate dehydrogenase 1 mutant inhibitor ivosidenib can be overcome by alternative dimer-interface binding inhibitors.
Nat Commun, 13, 2022
|
|
8DTX
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV89-22 | | Descriptor: | COV89-22 heavy chain, COV89-22 light chain, Spike protein S2' stem helix peptide | | Authors: | Lin, T.H, Lee, C.C.D, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
7PJN
 
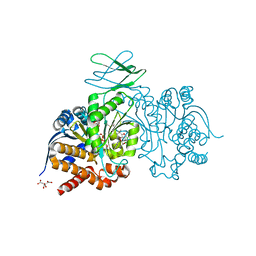 | | Crystal Structure of Ivosidenib-resistant IDH1 variant R132C S280F in complex with NADPH and inhibitor DS-1001B | | Descriptor: | (E)-3-(1-(5-(2-fluoropropan-2-yl)-3-(2,4,6-trichlorophenyl)isoxazole-4-carbonyl)-3-methyl-1H-indol-4-yl)acrylic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Reinbold, R, Rabe, P, Abboud, M.I, Schofield, C.J, Clifton, I.J. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Resistance to the isocitrate dehydrogenase 1 mutant inhibitor ivosidenib can be overcome by alternative dimer-interface binding inhibitors.
Nat Commun, 13, 2022
|
|
8P65
 
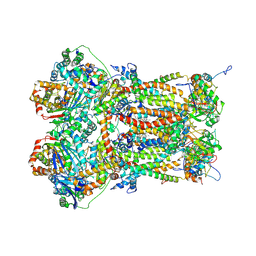 | | Cytochrome bc1 complex (Bos taurus) | | Descriptor: | Cytochrome b, Cytochrome b-c1 complex subunit 1, mitochondrial, ... | | Authors: | Phillips, B.P, Barra, I.M.C.C, Meier, T.K, Rimle, L, von Ballmoos, C. | | Deposit date: | 2023-05-25 | | Release date: | 2024-07-03 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A splendid molecular factory: De- and reconstruction of the mammalian respiratory chain.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
3D8L
 
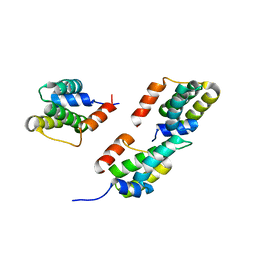 | | Crystal structure of ORF12 from the lactococcus lactis bacteriophage p2 | | Descriptor: | ORF12 | | Authors: | Siponen, M.I, Spinelli, S, Lichiere, J, Moineau, S, Cambillau, C, Campanacci, V. | | Deposit date: | 2008-05-23 | | Release date: | 2009-04-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of ORF12 from Lactococcus lactis phage p2 identifies a tape measure protein chaperone
J.Bacteriol., 191, 2009
|
|
3D8H
 
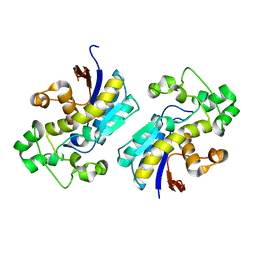 | | Crystal structure of phosphoglycerate mutase from Cryptosporidium parvum, cgd7_4270 | | Descriptor: | Glycolytic phosphoglycerate mutase | | Authors: | Wernimont, A.K, Lew, J, Wasney, G, Alam, Z, Kozieradzki, I, Cossar, D, Schapiro, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Wilkstrom, M, Edwards, A.M, Hui, R, Artz, J.D, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-23 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Characterization of a new phosphatase from Plasmodium.
Mol.Biochem.Parasitol., 179, 2011
|
|
6FL0
 
 | | Crystal structure of the membrane attack complex assembly inhibitor BGA71 from Lyme disease agent Borreliella bavariensis | | Descriptor: | membrane attack complex assembly inhibitor BGA71 | | Authors: | Brangulis, K, Akopjana, I, Petrovskis, I, Kazaks, A, Tars, K. | | Deposit date: | 2018-01-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the membrane attack complex assembly inhibitor BGA71 from the Lyme disease agent Borrelia bavariensis.
Sci Rep, 8, 2018
|
|
8WRC
 
 | |
7YKE
 
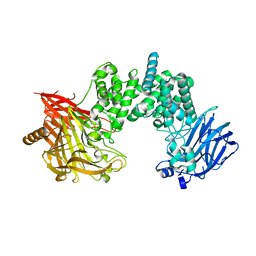 | | Crystal structure of chondroitin ABC lyase I in complex with chondroitin disaccharide 4,6-sulfate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4,6-di-O-sulfo-beta-D-galactopyranose, Chondroitin sulfate ABC endolyase, MAGNESIUM ION | | Authors: | Takashima, M, Watanabe, I, Miyanaga, A, Eguchi, T. | | Deposit date: | 2022-07-22 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Biochemical and crystallographic assessments of the effect of 4,6-O-disulfated disaccharide moieties in chondroitin sulfate E on chondroitinase ABC I activity.
Febs J., 290, 2023
|
|
7PI0
 
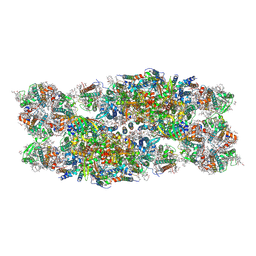 | | Unstacked compact Dunaliella PSII | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Caspy, I, Fadeeva, M, Mazor, Y, Nelson, N. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Structure of Dunaliella Photosystem II reveals conformational flexibility of stacked and unstacked supercomplexes.
Elife, 12, 2023
|
|
7PNK
 
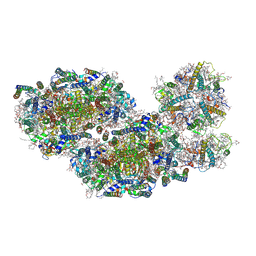 | | Unstacked compact Dunaliella PSII | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Caspy, I, Fadeeva, M, Mazor, Y, Nelson, N. | | Deposit date: | 2021-09-07 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structure of Dunaliella Photosystem II reveals conformational flexibility of stacked and unstacked supercomplexes.
Elife, 12, 2023
|
|
6FPJ
 
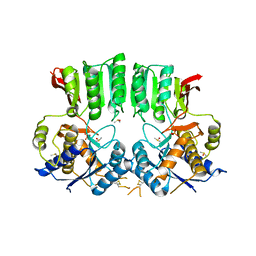 | | Structure of the AMPAR GluA3 N-terminal domain bound to phosphate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Herguedas, B, Garcia-Nafria, J, Greger, I. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Druggability Simulations and X-Ray Crystallography Reveal a Ligand-Binding Site in the GluA3 AMPA Receptor N-Terminal Domain.
Structure, 27, 2019
|
|
6FYT
 
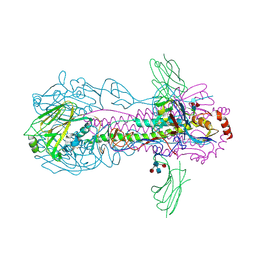 | |
8FX5
 
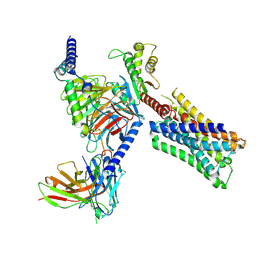 | | Human M4 muscarinic acetylcholine receptor complex with Gi1 and xanomeline | | Descriptor: | Antibody fragment scFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Vuckovic, Z, Mobbs, J.I, Glukhova, A, Sexton, P.M, Danev, R, Thal, D.M. | | Deposit date: | 2023-01-23 | | Release date: | 2023-09-13 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Xanomeline displays concomitant orthosteric and allosteric binding modes at the M 4 mAChR.
Nat Commun, 14, 2023
|
|
6TXA
 
 | | Crystal structure of tetrameric human D137N-SAMHD1 (residues 109-626) with XTP, dGMPNPP and Mg | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-13 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
6TXE
 
 | | Crystal structure of tetrameric human wt-SAMHD1 (residues 109-626) with GTP, dATP, dTMPNPP and Mg | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A.G, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-14 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
8K3Y
 
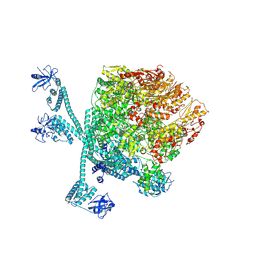 | | The "5+1" heteromeric structure of Lon protease consisting of a spiral pentamer with Y224S mutation and an N-terminal-truncated monomeric E613K mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Zhang, K, Chang, C.I. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
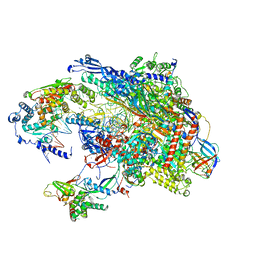
5VVR
 
 | |
