4CAV
 
 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a benzofuran ligand R0-09-4879 | | Descriptor: | 3-[[3-methyl-2-[[2,3,4-tris(fluoranyl)phenoxy]methyl]-1-benzofuran-4-yl]oxy]-N-(pyridin-3-ylmethyl)propan-1-amine, CHLORIDE ION, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Robinson, D.A, Fang, W, Raimi, O.G, Blair, D.E, Harrison, J, Ruda, G.F, Lockhart, D.E.A, Torrie, L.S, Wyatt, P.G, Gilbert, I.H, Van Aalten, D.M.F. | | Deposit date: | 2013-10-09 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | N-Myristoyltransferase is a Cell Wall Target in Aspergillus Fumigatus.
Acs Chem.Biol., 10, 2015
|
|
3ZJ0
 
 | | The human O-GlcNAcase C-terminal domain is a pseudo histone acetyltransferase | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, ACETYLTRANSFERASE, ... | | Authors: | Rao, F.V, Schuettelkopf, A.W, Dorfmueller, H.C, Ferenbach, A.T, Navratilova, I, van Aalten, D.M.F. | | Deposit date: | 2013-01-15 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Bacterial Putative Acetyltransferase Defines the Fold of the Human O-Glcnacase C-Terminal Domain.
Open Biol., 3, 2013
|
|
3ZZI
 
 | | Crystal structure of a tetrameric acetylglutamate kinase from Saccharomyces cerevisiae | | Descriptor: | ACETYLGLUTAMATE KINASE | | Authors: | de Cima, S, Gil-Ortiz, F, Crabeel, M, Fita, I, Rubio, V. | | Deposit date: | 2011-09-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Insight on an Arginine Synthesis Metabolon from the Tetrameric Structure of Yeast Acetylglutamate Kinase
Plos One, 7, 2012
|
|
3ZXP
 
 | |
4BVV
 
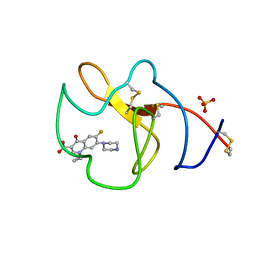 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, APOLIPOPROTEIN(A), SULFATE ION | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4C0W
 
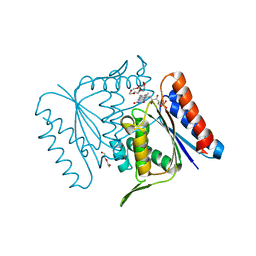 | |
4C14
 
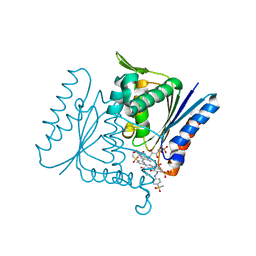 | | The crystal strucuture of PpAzoR in complex with reactive black 5 (RB5) | | Descriptor: | DODECAETHYLENE GLYCOL, FMN-DEPENDENT NADH-AZOREDUCTASE 1, [5-[3-[2-[[4-[2-[1-azanyl-7-[2-[4-[methyl-bis(oxidanyl)-$l^{4}-sulfanyl]phenyl]hydrazinyl]-8-oxidanyl-3,6-bis[tris(oxidanyl)-$l^{4}-sulfanyl]naphthalen-2-yl]hydrazinyl]phenyl]-bis(oxidanyl)-$l^{4}-sulfanyl]ethoxy]-7,8-dimethyl-2,4-bis(oxidanylidene)benzo[g]pteridin-10-yl]-2,3,4-tris(oxidanyl)pentyl] dihydrogen phosphate | | Authors: | Goncalves, A.M.D, de Sanctis, D, Bento, I. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of Pseudomonas putida azoreductase - the active site revisited.
FEBS J., 280, 2013
|
|
4BX4
 
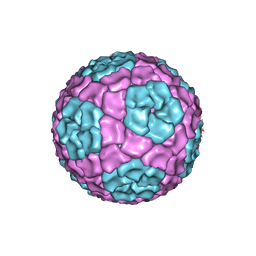 | | Fitting of the bacteriophage Phi8 P1 capsid protein into cryo-EM density | | Descriptor: | P1 | | Authors: | El Omari, K, Sutton, G, Ravantti, J.J, Zhang, H, Walter, T.S, Grimes, J.M, Bamford, D.H, Stuart, D.I, Mancini, E.J. | | Deposit date: | 2013-07-08 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Plate Tectonics of Virus Shell Assembly and Reorganization in Phage Phi8, a Distant Relative of Mammalian Reoviruses
Structure, 21, 2013
|
|
4CAY
 
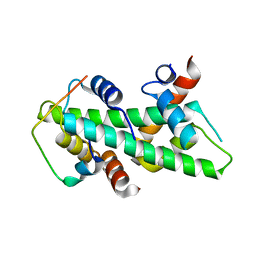 | | Crystal structure of a human Anp32e-H2A.Z-H2B complex | | Descriptor: | ACIDIC LEUCINE-RICH NUCLEAR PHOSPHOPROTEIN 32 FAMILY MEMBER E, HISTONE H2A.Z, HISTONE H2B TYPE 1-J | | Authors: | Obri, A, Ouararhni, K, Papin, C, Diebold, M.-L, Padmanabhan, K, Marek, M, Stoll, I, Roy, L, Reilly, P.T, Mak, T.W, Dimitrov, S, Romier, C, Hamiche, A. | | Deposit date: | 2013-10-09 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Anp32E is a Histone Chaperone that Removes H2A.Z from Chromatin
Nature, 648, 2014
|
|
4CFM
 
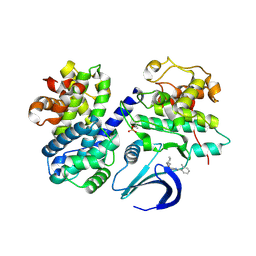 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 6-(cyclohexylmethoxy)-8-(2-methylphenyl)-9H-purin-2-amine, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-18 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4CBE
 
 | | Crystal structure of complement factors H and FHL-1 binding protein BBH06 or CRASP-2 from Borrelia burgdorferi (Native) | | Descriptor: | COMPLEMENT REGULATOR-ACQUIRING SURFACE PROTEIN 2 (CRASP-2) | | Authors: | Brangulis, K, Petrovskis, I, Kazaks, A, Ranka, R, Tars, K. | | Deposit date: | 2013-10-13 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Characterization of Cspz, a Complement Regulator Factor H and Fhl-1 Binding Protein from Borrelia Burgdorferi.
FEBS J., 281, 2014
|
|
4C47
 
 | |
3WRF
 
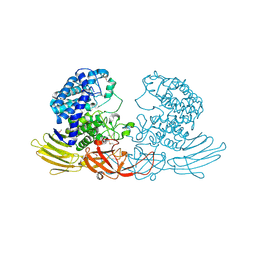 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
3WW0
 
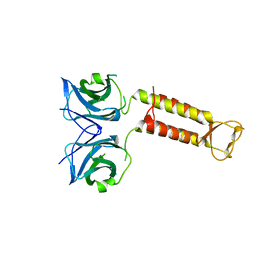 | | Crystal structure of F97A mutant, a new nuclear transport receptor of Hsp70 | | Descriptor: | Protein Hikeshi | | Authors: | Song, J, Kose, S, Watanabe, A, Son, S.Y, Choi, S, Hong, R.H, Yamashita, E, Park, I.Y, Imamoto, N, Lee, S.J. | | Deposit date: | 2014-06-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional analysis of Hikeshi, a new nuclear transport receptor of Hsp70s
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WBJ
 
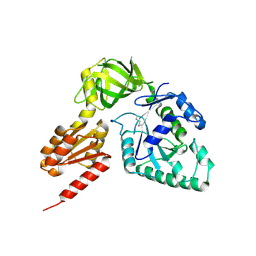 | | Crystal structure analysis of eukaryotic translation initiation factor 5B structure II | | Descriptor: | Eukaryotic translation initiation factor 5B | | Authors: | Zheng, A, Yamamoto, R, Ose, T, Yu, J, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-20 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | X-ray structures of eIF5B and the eIF5B-eIF1A complex: the conformational flexibility of eIF5B is restricted on the ribosome by interaction with eIF1A
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3X0L
 
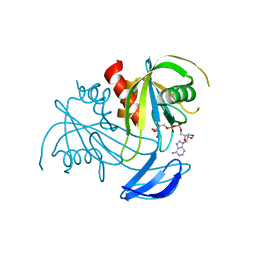 | | ADP ribose pyrophosphatase from Thermus thermophilus HB8 in ES-state at 1.00 angstrom resolution | | Descriptor: | ADP-ribose pyrophosphatase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-17 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
3X06
 
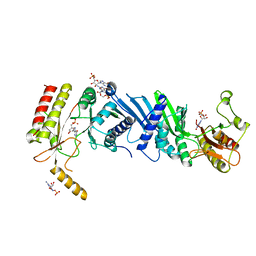 | | Crystal structure of PIP4KIIBETA T201M complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X0Q
 
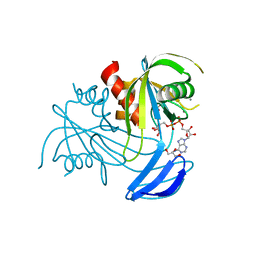 | | ADP ribose pyrophosphatase from Thermus thermophilus HB8 in ESMM-state at reaction time of 20 min | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-17 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
3X2O
 
 | | Neutron and X-ray joint refined structure of PcCel45A apo form at 298K. | | Descriptor: | Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Tanaka, I, Niimura, N, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2019-12-18 | | Method: | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X0K
 
 | | ADP ribose pyrophosphatase from Thermus thermophilus HB8 in ES-state at 0.97 angstrom resolution | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-16 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
4CAX
 
 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a pyrazole sulphonamide ligand (DDD85646) | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Raimi, O.G, Robinson, D.A, Fang, W, Blair, D.E, Harrison, J, Ruda, G.F, Lockhart, D.E.A, Torrie, L.S, Wyatt, P.G, Gilbert, I.H, Van Aalten, D.M.F. | | Deposit date: | 2013-10-09 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N-Myristoyltransferase is a Cell Wall Target in Aspergillus Fumigatus.
Acs Chem.Biol., 10, 2015
|
|
4CIU
 
 | | Crystal structure of E. coli ClpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHAPERONE PROTEIN CLPB | | Authors: | Kopp, J, Sinning, I, Bukau, B, Kummer, E, Mogk, A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-05-14 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Cooperation with Hsp70 in Protein Disaggregation
Elife, 3, 2014
|
|
4CG0
 
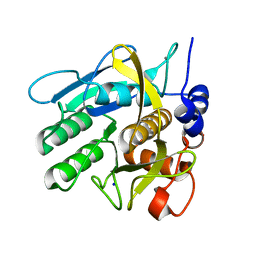 | | Savinase crystal structures for combined single crystal diffraction and powder diffraction analysis | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN SAVINASE | | Authors: | Frankaer, C.G, Moroz, O.V, Turkenburg, J.P, Aspmo, S.I, Thymark, M, Friis, E.P, Stahla, K, Nielsen, J.E, Wilson, K.S, Harris, P. | | Deposit date: | 2013-11-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Analysis of an Industrial Production Suspension of Bacillus Lentus Subtilisin Crystals by Powder Diffraction: A Powerful Quality-Control Tool.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CI5
 
 | |
4D2U
 
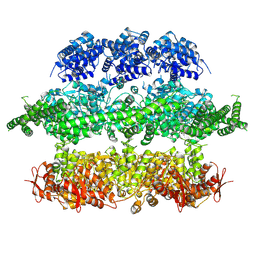 | | Negative-stain electron microscopy of E. coli ClpB (BAP form bound to ClpP) | | Descriptor: | CHAPERONE PROTEIN CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-04 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
