4W5R
 
 | |
7P77
 
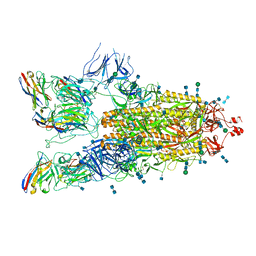 | | SARS-CoV-2 spike protein in complex with sybody#15 and sybody#68 in a 3up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
4V92
 
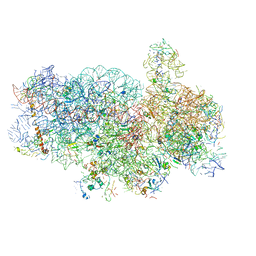 | | Kluyveromyces lactis 80S ribosome in complex with CrPV-IRES | | Descriptor: | 18S RRNA, ES1, ES10, ... | | Authors: | Fernandez, I.S, Bai, X, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-03-21 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Initiation of Translation by Cricket Paralysis Virus Ires Requires its Translocation in the Ribosome.
Cell(Cambridge,Mass.), 157, 2014
|
|
4W5Q
 
 | |
3KZW
 
 | | Crystal structure of cytosol aminopeptidase from Staphylococcus aureus COL | | Descriptor: | CHLORIDE ION, Cytosol aminopeptidase, PHOSPHATE ION, ... | | Authors: | Hattne, J, Dubrovska, I, Halavaty, A, Minasov, G, Scott, P, Shuvalova, L, Winsor, J, Otwinowski, Z, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-08 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: |
|
|
5LOK
 
 | | X-ray structure of uridine phosphorylase from Vibrio cholerae in complex with cytidine and cytosine at 1.11 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, 6-AMINOPYRIMIDIN-2(1H)-ONE, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Dontsova, M.V, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2016-08-09 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.109 Å) | | Cite: | X-ray structure of uridine phosphorylase from Vibrio cholerae in complex with cytidine and cytosine at 1.11 A resolution
To Be Published
|
|
6OCB
 
 | |
3KPD
 
 | | Crystal Structure of the CBS domain pair of protein MJ0100 in complex with 5 -methylthioadenosine and S-adenosyl-L-methionine. | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, S-ADENOSYLMETHIONINE, Uncharacterized protein MJ0100 | | Authors: | Lucas, M, Oyenarte, I, Garcia, I.G, Arribas, E.A, Encinar, J.A, Kortazar, D, Fernandez, J.A, Mato, J.M, Martinez-Chantar, M.L, Martinez-Cruz, L.A. | | Deposit date: | 2009-11-16 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Binding of S-Methyl-5'-Thioadenosine and S-Adenosyl-l-Methionine to Protein MJ0100 Triggers an Open-to-Closed Conformational Change in Its CBS Motif Pair.
J.Mol.Biol., 396, 2010
|
|
6OO2
 
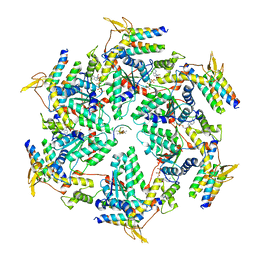 | | Vps4 with Cyclic Peptide Bound in the Central Pore | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Designed Cyclic Peptide, ... | | Authors: | Han, H, Fulcher, J.M, Dandey, V.P, Sundquist, W.I, Kay, M.S, Shen, P, Hill, C.P. | | Deposit date: | 2019-04-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of Vps4 with circular peptides and implications for translocation of two polypeptide chains by AAA+ ATPases.
Elife, 8, 2019
|
|
3KPB
 
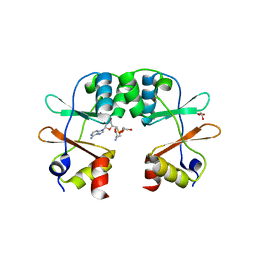 | | Crystal Structure of the CBS domain pair of protein MJ0100 in complex with 5 -methylthioadenosine and S-adenosyl-L-methionine. | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, Uncharacterized protein MJ0100 | | Authors: | Lucas, M, Oyenarte, I, Garcia, I.G, Arribas, E.A, Encinar, J.A, Kortazar, D, Fernandez, J.A, Mato, J.M, Martinez-Cruz, L.A. | | Deposit date: | 2009-11-16 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding of S-Methyl-5'-Thioadenosine and S-Adenosyl-l-Methionine to Protein MJ0100 Triggers an Open-to-Closed Conformational Change in Its CBS Motif Pair.
J.Mol.Biol., 396, 2010
|
|
3TG0
 
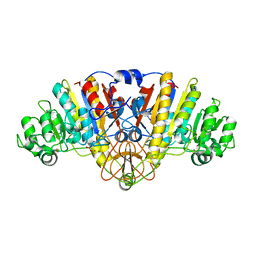 | | E. coli alkaline phosphatase with bound inorganic phosphate | | Descriptor: | Alkaline phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Bobyr, E, Lassila, J.K, Wiersma-Koch, H.I, Fenn, T.D, Lee, J.J, Nikolic-Hughes, I, Hodgson, K.O, Rees, D.C, Hedman, B, Herschlag, D. | | Deposit date: | 2011-08-16 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution analysis of Zn(2+) coordination in the alkaline phosphatase superfamily by EXAFS and x-ray crystallography.
J.Mol.Biol., 415, 2012
|
|
4WQC
 
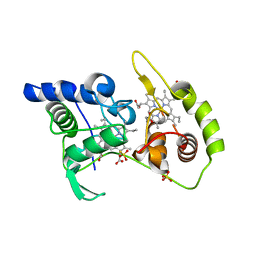 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - K208N mutant | | Descriptor: | 1,2-ETHANEDIOL, HEME C, SULFATE ION, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
1VTX
 
 | |
4WY2
 
 | | Crystal structure of universal stress protein E from Proteus mirabilis in complex with UDP-3-O-[(3R)-3-hydroxytetradecanoyl]-N-acetyl-alpha-glucosamine | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Shumilin, I.A, Shabalin, I.G, Handing, K.B, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-15 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of universal stress protein E from Proteus mirabilis incomplex withUDP-3-O-[(3R)-3-hydroxytetradecanoyl]-N-acetyl-alpha-glucosamine
to be published
|
|
6DHH
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.2 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
1W8X
 
 | | Structural analysis of PRD1 | | Descriptor: | MAJOR CAPSID PROTEIN (PROTEIN P3), PROTEIN P16, PROTEIN P30, ... | | Authors: | Abrescia, N.G.A, Cockburn, J.J.B, Grimes, J.M, Sutton, G.C, Diprose, J.M, Butcher, S.J, Fuller, S.D, San Martin, C, Burnett, R.M, Stuart, D.I, Bamford, D.H, Bamford, J.K.H. | | Deposit date: | 2004-10-01 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Insights Into Assembly from Structural Analysis of Bacteriophage Prd1.
Nature, 432, 2004
|
|
1VZY
 
 | | Crystal structure of the Bacillus subtilis HSP33 | | Descriptor: | 33 KDA CHAPERONIN, ACETATE ION, ZINC ION | | Authors: | Janda, I.K, Devedjiev, Y, Derewenda, U, Dauter, Z, Bielnicki, J, Cooper, D.R, Joachimiak, A, Derewenda, Z.S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-29 | | Release date: | 2004-10-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The crystal structure of the reduced, Zn2+-bound form of the B. subtilis Hsp33 chaperone and its implications for the activation mechanism.
Structure, 12, 2004
|
|
4E2S
 
 | |
1W76
 
 | | Orthorhombic form of Torpedo californica acetylcholinesterase (AChE) complexed with bis-acting galanthamine derivative | | Descriptor: | (-)-GALANTHAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE | | Authors: | Greenblatt, H.M, Guillou, C, Guenard, D, Badet, B, Thal, C, Silman, I, Sussman, J.L. | | Deposit date: | 2004-08-30 | | Release date: | 2004-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The complex of a bivalent derivative of galanthamine with torpedo acetylcholinesterase displays drastic deformation of the active-site gorge: implications for structure-based drug design.
J.Am.Chem.Soc., 126, 2004
|
|
2FX9
 
 | | Crystal structure of hiv-1 neutralizing human fab 4e10 in complex with a thioether-linked peptide encompassing the 4e10 epitope on gp41 | | Descriptor: | Fab 4E10, Fragment of HIV glycoprotein gp41 | | Authors: | Cardoso, R.M.F, Brunel, F.M, Ferguson, S, Burton, D.R, Dawson, P.E, Wilson, I.A. | | Deposit date: | 2006-02-03 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10.
J.Mol.Biol., 365, 2007
|
|
4E6K
 
 | | 2.0 A resolution structure of Pseudomonas aeruginosa bacterioferritin (BfrB) in complex with bacterioferritin associated ferredoxin (Bfd) | | Descriptor: | Bacterioferritin, FE2/S2 (INORGANIC) CLUSTER, PHOSPHATE ION, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Wang, Y, Kumar, R, Ruvinsky, A, Vasker, I, Rivera, M. | | Deposit date: | 2012-03-15 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of the BfrB-Bfd Complex Reveals Protein-Protein Interactions Enabling Iron Release from Bacterioferritin.
J.Am.Chem.Soc., 134, 2012
|
|
1W4B
 
 | | P4 protein from PHI12 in complex with product (AMPcPP Mg 22C) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1W46
 
 | | P4 protein from Bacteriophage PHI12 in complex with ADP and MG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
2FX8
 
 | | Crystal structure of hiv-1 neutralizing human fab 4e10 in complex with an aib-induced peptide encompassing the 4e10 epitope on gp41 | | Descriptor: | Fab 4E10, Fragment of HIV glycoprotein (GP41) | | Authors: | Cardoso, R.M.F, Brunel, F.M, Ferguson, S, Burton, D.R, Dawson, P.E, Wilson, I.A. | | Deposit date: | 2006-02-03 | | Release date: | 2006-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10.
J.Mol.Biol., 365, 2007
|
|
2FZV
 
 | | Crystal Structure of an apo form of a Flavin-binding Protein from Shigella flexneri | | Descriptor: | CALCIUM ION, CHLORIDE ION, putative arsenical resistance protein | | Authors: | Vorontsov, I.I, Minasov, G, Brunzelle, J.S, Shuvalova, L, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-10 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an apo form of Shigella flexneri ArsH protein with an NADPH-dependent FMN reductase activity
Protein Sci., 16, 2007
|
|
