2M5J
 
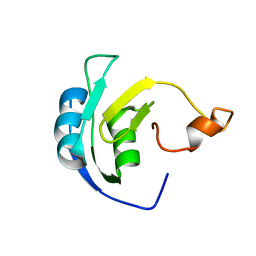 | | Solution structure of the periplasmic signaling domain of HasR, a TonB-dependent outer membrane heme transporter | | Descriptor: | HasR protein | | Authors: | Malki, I, Cardoso de Amorim, G, Prochnicka-Chalufour, A, Simenel, C, Delepierre, M, Izadi-Pruneyre, N. | | Deposit date: | 2013-02-26 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interaction of a Partially Disordered Antisigma Factor with Its Partner, the Signaling Domain of the TonB-Dependent Transporter HasR
Plos One, 9, 2014
|
|
3LZF
 
 | |
5M7T
 
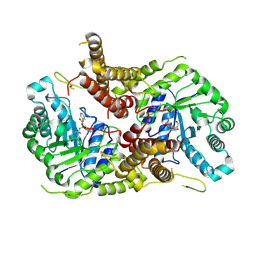 | | Structure of human O-GlcNAc hydrolase with PugNAc type inhibitor | | Descriptor: | (5R,6R,7R,8S)-8-(ACETYLAMINO)-6,7-DIHYDROXY-5-(HYDROXYMETHYL)-N-PHENYL-1,5,6,7,8,8A-HEXAHYDROIMIDAZO[1,2-A]PYRIDINE-2-CARBOXAMIDE, Protein O-GlcNAcase | | Authors: | Roth, C, Chan, S, Offen, W.A, Hemsworth, G.R, Willems, L.I, King, D, Varghese, V, Britton, R, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-29 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional insight into human O-GlcNAcase.
Nat. Chem. Biol., 13, 2017
|
|
5M8K
 
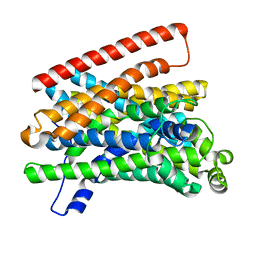 | | Crystal structure of Eremococcus coleocola manganese transporter mutant E129Q | | Descriptor: | Divalent metal cation transporter MntH | | Authors: | Manatschal, C, Ehrnstorfer, I.A, Arnold, F.M, Laederach, J, Dutzler, R. | | Deposit date: | 2016-10-29 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and mechanistic basis of proton-coupled metal ion transport in the SLC11/NRAMP family.
Nat Commun, 8, 2017
|
|
5MBJ
 
 | | Structure of a bacterial light-regulated adenylyl cyclase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, FLAVIN MONONUCLEOTIDE | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
2MOL
 
 | | 3D NMR structure of the cytoplasmic rhodanese domain of the full-length inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | Deposit date: | 2014-04-27 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
5MBE
 
 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, FLAVIN MONONUCLEOTIDE | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
1CDU
 
 | | STRUCTURE OF T-CELL SURFACE GLYCOPROTEIN CD4 MUTANT WITH PHE 43 REPLACED BY VAL | | Descriptor: | T-CELL SURFACE GLYCOPROTEIN CD4 | | Authors: | Wu, H, Myszka, D, Tendian, S.W, Brouillette, C.G, Sweet, R.W, Chaiken, I.M, Hendrickson, W.A. | | Deposit date: | 1996-11-11 | | Release date: | 1997-04-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Kinetic and structural analysis of mutant CD4 receptors that are defective in HIV gp120 binding.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1EGG
 
 | | STRUCTURE OF A C-TYPE CARBOHYDRATE-RECOGNITION DOMAIN (CRD-4) FROM THE MACROPHAGE MANNOSE RECEPTOR | | Descriptor: | CALCIUM ION, MACROPHAGE MANNOSE RECEPTOR | | Authors: | Feinberg, H, Park-Snyder, S, Kolatkar, A.R, Heise, C.T, Taylor, M.E, Weis, W.I. | | Deposit date: | 2000-02-15 | | Release date: | 2000-08-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a C-type carbohydrate recognition domain from the macrophage mannose receptor.
J.Biol.Chem., 275, 2000
|
|
4YDX
 
 | | Crystal structure of cisplatin bound to a human copper chaperone (monomer) - new refinement | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Copper transport protein ATOX1, PLATINUM (II) ION, ... | | Authors: | Shabalin, I.G, Boal, A.K, Dauter, Z, Jaskolski, M, Minor, W, Rosenzweig, A.C, Wlodawer, A. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YEA
 
 | | Crystal structure of cisplatin bound to a human copper chaperone (dimer) - new refinement | | Descriptor: | COPPER (II) ION, Copper transport protein ATOX1, SULFATE ION | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1EHW
 
 | | HUMAN NUCLEOSIDE DIPHOSPHATE KINASE 4 | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE, SULFATE ION | | Authors: | Milon, L, Meyer, P, Chiadmi, M, Munier, A, Johansson, M, Karlsson, A, Lascu, I, Capeau, J, Janin, J, Lacombe, M.-L. | | Deposit date: | 2000-02-23 | | Release date: | 2000-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase.
J.Biol.Chem., 275, 2000
|
|
2MRE
 
 | | NMR structure of the Rad18-UBZ/ubiquitin complex | | Descriptor: | E3 ubiquitin-protein ligase RAD18, Polyubiquitin-C, ZINC ION | | Authors: | Rizzo, A.A, Salerno, P.E, Bezsonova, I, Korzhnev, D.M. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Human Rad18 Zinc Finger in Complex with Ubiquitin Defines a Class of UBZ Domains in Proteins Linked to the DNA Damage Response.
Biochemistry, 53, 2014
|
|
5M7S
 
 | | Structure of human O-GlcNAc hydrolase with bound transition state analog ThiametG | | Descriptor: | (3AR,5R,6S,7R,7AR)-2-(ETHYLAMINO)-5-(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]THIAZOLE-6,7-DIOL, Protein O-GlcNAcase | | Authors: | Roth, C, Chan, S, Offen, W.A, Hemsworth, G.R, Willems, L.I, King, D, Varghese, V, Britton, R, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-29 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insight into human O-GlcNAcase.
Nat. Chem. Biol., 13, 2017
|
|
2VA9
 
 | | Structure of native TcAChE after a 9 seconds annealing to room temperature during the first 5 seconds of which laser irradiation at 266nm took place | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE | | Authors: | Colletier, J.-P, Sanson, B, Royant, A, Specht, A, Nachon, F, Masson, P, Zaccai, G, Sussman, J.L, Goeldner, M, Silman, I, Bourgeois, D, Weik, M. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Use of a 'Caged' Analog to Study Traffic of Choline within Acetylcholinesterase by Kinetic Crystallography
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5MI0
 
 | | A thermally stabilised version of Plasmodium falciparum RH5 | | Descriptor: | MONOCLONAL ANTIBODY 9AD4, Reticulocyte binding-like protein 5,Reticulocyte binding protein 5 | | Authors: | Campeotto, I, Goldenzweig, A, Davey, J, Barfod, L, Marshall, J.M, Silk, S.E, Wright, K.E, Draper, S.J, Higgins, M.K, Fleishman, S.J. | | Deposit date: | 2016-11-27 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | One-step design of a stable variant of the malaria invasion protein RH5 for use as a vaccine immunogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MBH
 
 | | Structure of a bacterial light-regulated adenylyl cyclase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, FLAVIN MONONUCLEOTIDE | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
5MHQ
 
 | | CCT068127 in complex with CDK2 | | Descriptor: | (2~{R},3~{S})-3-[[9-propan-2-yl-6-(pyridin-3-ylmethylamino)purin-2-yl]amino]pentan-2-ol, Cyclin-dependent kinase 2 | | Authors: | Whittaker, S.R, Barlow, C, Martin, M.P, Mancusi, C, Wagner, S, Barrie, E, te Poele, R, Sharp, S, Brown, N, Wilson, S, Clarke, P, Walton, M.I, MacDonald, E, Blagg, J, Noble, M.E.M, Garrett, M.D, Workman, P. | | Deposit date: | 2016-11-25 | | Release date: | 2017-12-20 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular profiling and combinatorial activity of CCT068127: a potent CDK2 and CDK9 inhibitor.
Mol Oncol, 12, 2018
|
|
2N3Y
 
 | | NMR structure of the Y48pCMF variant of human cytochrome c in its reduced state | | Descriptor: | Cytochrome c, Mesoheme | | Authors: | Moreno-Beltran, B, Del Conte, R, Diaz-Quintana, A, De la Rosa, M.A, Turano, P, Diaz-Moreno, I. | | Deposit date: | 2015-06-15 | | Release date: | 2016-12-14 | | Last modified: | 2017-04-26 | | Method: | SOLUTION NMR | | Cite: | Structural basis of mitochondrial dysfunction in response to cytochrome c phosphorylation at tyrosine 48.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2N2H
 
 | | Solution structure of Sds3 in complex with Sin3A | | Descriptor: | Paired amphipathic helix protein Sin3a, Sin3 histone deacetylase corepressor complex component SDS3 | | Authors: | Clark, M, Radhakrishnan, I. | | Deposit date: | 2015-05-08 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the assembly of the histone deacetylase-associated Sin3L/Rpd3L corepressor complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2V14
 
 | | Kinesin 16B Phox-homology domain (KIF16B) | | Descriptor: | KINESIN-LIKE MOTOR PROTEIN C20ORF23 | | Authors: | Wilson, M.I, Williams, R.L, Cho, W, Hong, W, Blatner, N.R. | | Deposit date: | 2007-05-21 | | Release date: | 2007-07-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis of Novel Endosome Anchoring Activity of Kif16B Kinesin.
Embo J., 26, 2007
|
|
2MZ3
 
 | |
4ZO1
 
 | | Crystal Structure of the T3-bound TR-beta Ligand-binding Domain in complex with RXR-alpha | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha, ... | | Authors: | Bruning, J.B, Kojetin, D.J, Matta-Camacho, E, Hughes, T.S, Srinivasan, S, Nwachukwu, J.C, Cavett, V, Nowak, J, Chalmers, M.J, Marciano, D.P, Kamenecka, T.M, Rance, M, Shulman, A.I, Mangelsdorf, D.J, Griffin, P.R, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.221 Å) | | Cite: | Structural mechanism for signal transduction in RXR nuclear receptor heterodimers.
Nat Commun, 6, 2015
|
|
3T6X
 
 | | Crystal Structure of Steccherinum ochraceum Laccase obtained by multi-crystals composite data collection technique (20% dose) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I, Kolomytseva, M, Golovleva, L, Scozzafava, A, Chernykh, A.M. | | Deposit date: | 2011-07-29 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Reaction intermediates and redox state changes in a blue laccase from Steccherinum ochraceum observed by crystallographic high/low X-ray dose experiments.
J.Inorg.Biochem., 111, 2012
|
|
5O8R
 
 | | The crystal structure of DfoA bound to FAD and NADP; the desferrioxamine biosynthetic pathway cadaverine monooxygenase from the fire blight disease pathogen Erwinia amylovora | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-lysine 6-monooxygenase involved in desferrioxamine biosynthesis, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | Deposit date: | 2017-06-14 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J. Struct. Biol., 202, 2018
|
|
