5JOM
 
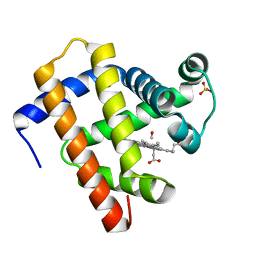 | | X-ray structure of CO-bound sperm whale myoglobin using a fixed target crystallography chip | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Oghbaey, S, Sarracini, A, Ginn, H.M, Pare-Labrosse, O, Kuo, A, Marx, A, Epp, S.W, Sherrell, D.A, Eger, B.T, Zhong, Y, Loch, R, Mariani, V, Alonso-Mori, R, Nelson, S, Lemke, H.T, Owen, R.L, Pearson, A.R, Stuart, D.I, Ernst, O.P, Mueller-Werkmeister, H.M, Miller, R.J.D. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fixed target combined with spectral mapping: approaching 100% hit rates for serial crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5JQY
 
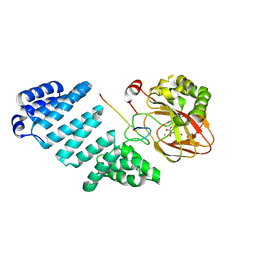 | |
5JPZ
 
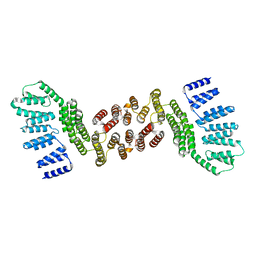 | | Crystal structure of HAT domain of human Squamous Cell Carcinoma Antigen Recognized By T Cells 3, SART3 (TIP110) | | Descriptor: | Squamous cell carcinoma antigen recognized by T-cells 3 | | Authors: | Grazette, A, Harper, S, Emsley, J, Layfield, R, Dreveny, I. | | Deposit date: | 2016-05-04 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.045 Å) | | Cite: | unpublished
To Be Published
|
|
5JQH
 
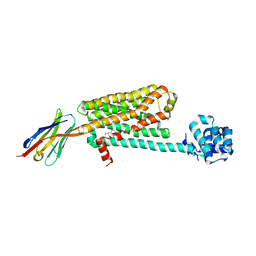 | | Structure of beta2 adrenoceptor bound to carazolol and inactive-state stabilizing nanobody, Nb60 | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, CHOLESTEROL, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Staus, D.P, Strachan, R.T, Manglik, A, Pani, B, Kahsai, A.W, Kim, T.H, Wingler, L.M, Ahn, S, Chatterjee, A, Masoudi, A, Kruse, A.C, Pardon, E, Steyaert, J, Weis, W.I, Prosser, R.S, Kobilka, B.K, Costa, T, Lefkowitz, R.J. | | Deposit date: | 2016-05-05 | | Release date: | 2016-07-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation.
Nature, 535, 2016
|
|
5JUB
 
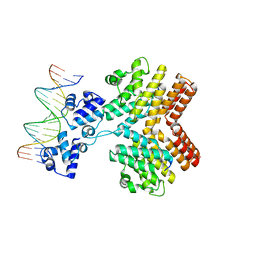 | | Crystal structure of ComR from S.thermophilus in complex with DNA and its signalling peptide ComS. | | Descriptor: | ComS, Transcriptional regulator, pComX-for, ... | | Authors: | Talagas, A, Fontaine, L, Ledesma-Garcia, L, Li de la Sierra-Gallay, I, Hols, P, Nessler, S. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural Insights into Streptococcal Competence Regulation by the Cell-to-Cell Communication System ComRS.
PLoS Pathog., 12, 2016
|
|
5JUF
 
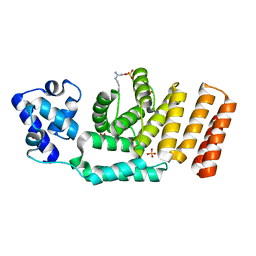 | | Crystal structure of the apo form of ComR from S. thermophilus. | | Descriptor: | SULFATE ION, Transcriptional regulator | | Authors: | Talagas, A, Fontaine, L, Ledesma, L, Li de la Sierra-Gallay, I, Hols, P, Nessler, S. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-26 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Structural Insights into Streptococcal Competence Regulation by the Cell-to-Cell Communication System ComRS.
PLoS Pathog., 12, 2016
|
|
6KYD
 
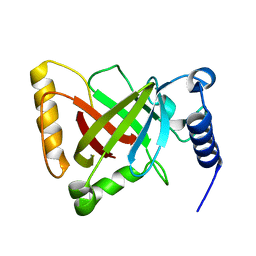 | | Structure of the R217A mutant of Clostridium difficile sortase B | | Descriptor: | Putative peptidase C60B, sortase B | | Authors: | Kang, C.Y, Huang, I.H, Wu, T.Y, Chang, J.C, Hsiao, Y.Y, Cheng, C.H, Tsai, W.J, Hsu, K.C, Wang, S.Y. | | Deposit date: | 2019-09-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Functional analysis ofClostridium difficilesortase B reveals key residues for catalytic activity and substrate specificity.
J.Biol.Chem., 295, 2020
|
|
2I25
 
 | |
5JZA
 
 | |
5JZU
 
 | |
2HQI
 
 | | NMR SOLUTION STRUCTURE OF THE OXIDIZED FORM OF MERP, 14 STRUCTURES | | Descriptor: | MERCURIC TRANSPORT PROTEIN | | Authors: | Qian, H, Sahlman, L, Eriksson, P.O, Hambreus, C, Edlund, U, Sethson, I. | | Deposit date: | 1998-03-31 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the oxidized form of MerP, a mercuric ion binding protein involved in bacterial mercuric ion resistance.
Biochemistry, 37, 1998
|
|
5JS2
 
 | | Human Argonaute-2 Bound to a Modified siRNA | | Descriptor: | MAGNESIUM ION, PHENOL, PHOSPHATE ION, ... | | Authors: | Schirle, N.T, MacRae, I.J. | | Deposit date: | 2016-05-07 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural Analysis of Human Argonaute-2 Bound to a Modified siRNA Guide.
J.Am.Chem.Soc., 138, 2016
|
|
6Z06
 
 | | Crystal structure of Puumala virus Gc in complex with Fab 4G2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope polyprotein, Fab 4G2 Heavy chain, ... | | Authors: | Rissanen, I.R, Stass, R, Krumm, S.A, Seow, J, Hulswit, R.J.G, Paesen, G.C, Hepojoki, J, Vapalahti, O, Lundkvist, A, Reynard, O, Volchkov, V, Doores, K.J, Huiskonen, J.T, Bowden, T.A. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular rationale for antibody-mediated targeting of the hantavirus fusion glycoprotein.
Elife, 9, 2020
|
|
2IDZ
 
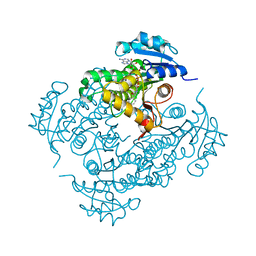 | | Crystal structure of wild type Enoyl-ACP(CoA) reductase from Mycobacterium tuberculosis in complex with NADH-INH | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], ISONICOTINIC-ACETYL-NICOTINAMIDE-ADENINE DINUCLEOTIDE | | Authors: | Dias, M.V.B, Prado, M.P.X, Vasconcelos, I.B, Valmir, F, Basso, L.A, Santos, D.S, Azevedo Jr, W.F. | | Deposit date: | 2006-09-15 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies on the binding of isonicotinyl-NAD adduct to wild-type and isoniazid resistant 2-trans-enoyl-ACP (CoA) reductase from Mycobacterium tuberculosis.
J.Struct.Biol., 159, 2007
|
|
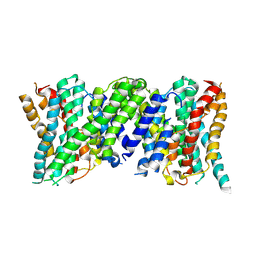
6Z3Y
 
 | |
8AFK
 
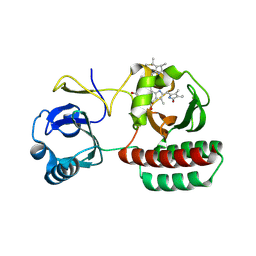 | | Structure of iRFP variant C15S/N136R/V256C in complex with phycocyanobilin | | Descriptor: | Near-infrared fluorescent protein, PHYCOCYANOBILIN | | Authors: | Remeeva, A, Kovalev, K, Gushchin, I, Fonin, A, Turoverov, K, Stepanenko, O. | | Deposit date: | 2022-07-18 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Structure of iRFP variant C15S/N136R/V256C in complex with phycocyanobilin
To Be Published
|
|
2IT6
 
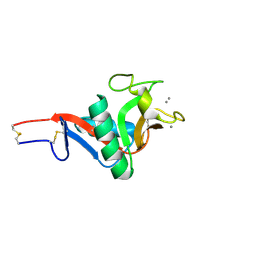 | | Crystal Structure of DCSIGN-CRD with man2 | | Descriptor: | CALCIUM ION, CD209 antigen, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Weis, W.I, Feinberg, H, Castelli, R, Drickamer, K, Seeberger, P.H. | | Deposit date: | 2006-10-19 | | Release date: | 2006-12-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Multiple modes of binding enhance the affinity of DC-SIGN for high mannose N-linked glycans found on viral glycoproteins.
J.Biol.Chem., 282, 2007
|
|
2IKO
 
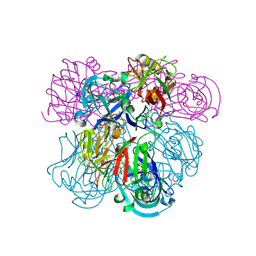 | | Crystal Structure of Human Renin Complexed with Inhibitor | | Descriptor: | 5-{4-[(3,5-DIFLUOROBENZYL)AMINO]PHENYL}-6-ETHYLPYRIMIDINE-2,4-DIAMINE, Renin | | Authors: | Mochalkin, I. | | Deposit date: | 2006-10-02 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding thermodynamics of substituted diaminopyrimidine renin inhibitors.
Anal.Biochem., 360, 2007
|
|
6YUX
 
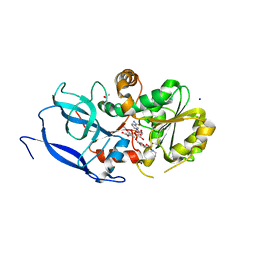 | | Crystal structure of Malus domestica Double Bond Reductase (MdDBR) ternary complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ... | | Authors: | Caliandro, R, Polsinelli, I, Demitri, N, Benini, S. | | Deposit date: | 2020-04-27 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The structural and functional characterization of Malus domestica double bond reductase MdDBR provides insights towards the identification of its substrates.
Int.J.Biol.Macromol., 171, 2021
|
|
2J5D
 
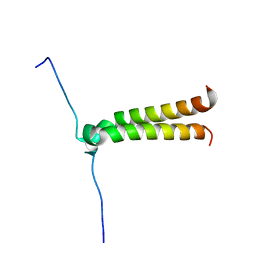 | | NMR structure of BNIP3 transmembrane domain in lipid bicelles | | Descriptor: | BCL2/ADENOVIRUS E1B 19 KDA PROTEIN-INTERACTING PROTEIN 3 | | Authors: | Bocharov, E.V, Pustovalova, Y.E, Volynsky, P.E, Maslennikov, I.V, Goncharuk, M.V, Ermolyuk, Y.S, Arseniev, A.S. | | Deposit date: | 2006-09-14 | | Release date: | 2007-04-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unique dimeric structure of BNip3 transmembrane domain suggests membrane permeabilization as a cell death trigger.
J. Biol. Chem., 282, 2007
|
|
5JXK
 
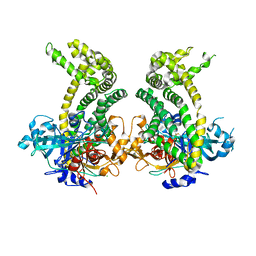 | | Crystal structure of Porphyromonas endodontalis DPP11 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5JZ8
 
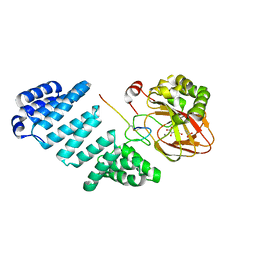 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH)oxygenase and TPR domains in complex with manganese, N-oxalylglycine, and factor X substrate peptide fragment (39mer) | | Descriptor: | ACETATE ION, Aspartyl/asparaginyl beta-hydroxylase, Coagulation factor X, ... | | Authors: | McDonough, M.A, Pfeffer, I. | | Deposit date: | 2016-05-16 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Aspartate/asparagine-beta-hydroxylase crystal structures reveal an unexpected epidermal growth factor-like domain substrate disulfide pattern.
Nat Commun, 10, 2019
|
|
5K24
 
 | |
6Z2L
 
 | |
5KI6
 
 | |
