3ROG
 
 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Thymidine 3'-monophosphate | | Descriptor: | Apolipoprotein A-I-binding protein, SULFATE ION, THYMIDINE-3'-PHOSPHATE | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-25 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
1Z68
 
 | | Crystal Structure Of Human Fibroblast Activation Protein alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aertgeerts, K, Levin, I, Shi, L, Prasad, G.S, Zhang, Y, Kraus, M.L, Salakian, S, Snell, G.P, Sridhar, V, Wijnands, R, Tennant, M.G. | | Deposit date: | 2005-03-21 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and kinetic analysis of the substrate specificity of human fibroblast activation protein alpha.
J.Biol.Chem., 280, 2005
|
|
6N4X
 
 | | Metabotropic Glutamate Receptor 5 Apo Form Ligand Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Metabotropic glutamate receptor 5 | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
1TP9
 
 | | PRX D (type II) from Populus tremula | | Descriptor: | SULFATE ION, peroxiredoxin | | Authors: | Echalier, A, Trivelli, X, Corbier, C, Rouhier, N, Walker, O, Tsan, P, Jacquot, J.P, Krimm, I, Lancelin, J.M. | | Deposit date: | 2004-06-16 | | Release date: | 2005-04-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure and solution NMR dynamics of a D (type II) peroxiredoxin glutaredoxin and thioredoxin dependent: a new insight into the peroxiredoxin oligomerism
Biochemistry, 44, 2005
|
|
3RNO
 
 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with NADP. | | Descriptor: | Apolipoprotein A-I-binding protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-22 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3ROZ
 
 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Nicotinamide | | Descriptor: | Apolipoprotein A-I-binding protein, NICOTINAMIDE, SULFATE ION | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
6MLY
 
 | | Bifunctional GH43-CE Bacteroides eggerthii, BACEGG_01304 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Bifunctional GH43-CE protein, ... | | Authors: | Koropatkin, N.M, Pereira, G.V, Cann, I. | | Deposit date: | 2018-09-28 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Degradation of complex arabinoxylans by human colonic Bacteroidetes
Nat Commun, 2021
|
|
2JJS
 
 | | Structure of human CD47 in complex with human signal regulatory protein (SIRP) alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IODIDE ION, LEUKOCYTE SURFACE ANTIGEN CD47, ... | | Authors: | Hatherley, D, Graham, S.C, Turner, J, Harlos, K, Stuart, D.I, Barclay, A.N. | | Deposit date: | 2008-04-22 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Paired receptor specificity explained by structures of signal regulatory proteins alone and complexed with CD47.
Mol. Cell, 31, 2008
|
|
1TXT
 
 | | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase | | Descriptor: | 3-hydroxy-3-methylglutaryl-CoA synthase, ACETOACETYL-COENZYME A | | Authors: | Campobasso, N, Patel, M, Wilding, I.E, Kallender, H, Rosenberg, M, Gwynn, M. | | Deposit date: | 2004-07-06 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase: crystal structure and mechanism
J.Biol.Chem., 279, 2004
|
|
6N2V
 
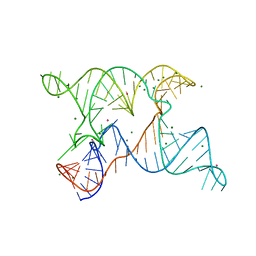 | |
1TP7
 
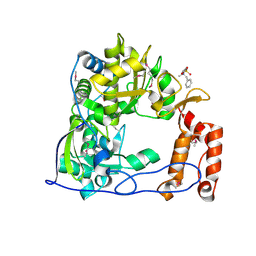 | | Crystal Structure of the RNA-dependent RNA Polymerase from Human Rhinovirus 16 | | Descriptor: | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, Genome polyprotein, SULFATE ION | | Authors: | Appleby, T.C, Luecke, H, Shim, J.H, Wu, J.Z, Cheney, I.W, Zhong, W, Vogeley, L, Hong, Z, Yao, N. | | Deposit date: | 2004-06-15 | | Release date: | 2005-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of complete rhinovirus RNA polymerase suggests front loading of protein primer.
J.Virol., 79, 2005
|
|
4QY9
 
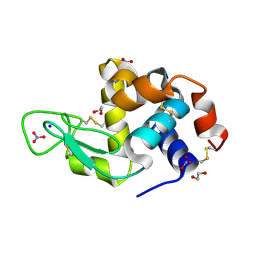 | | X-ray structure of the adduct between hen egg white lysozyme and Auoxo3, a cytotoxic gold(III) compound | | Descriptor: | 1,2-ETHANEDIOL, GOLD ION, Lysozyme C, ... | | Authors: | Russo Krauss, I, Merlino, A. | | Deposit date: | 2014-07-24 | | Release date: | 2014-11-05 | | Last modified: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Interactions of gold-based drugs with proteins: the structure and stability of the adduct formed in the reaction between lysozyme and the cytotoxic gold(iii) compound Auoxo3.
Dalton Trans, 43, 2014
|
|
1V0R
 
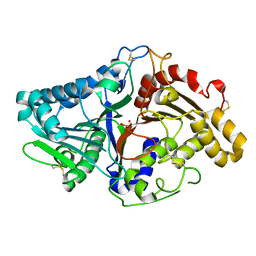 | |
3NS0
 
 | | X-ray structure of bacteriorhodopsin | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Borshchevskiy, V.I. | | Deposit date: | 2010-07-01 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-ray-Radiation-Induced Changes in Bacteriorhodopsin Structure.
J.Mol.Biol., 409, 2011
|
|
1V0V
 
 | |
3J5M
 
 | | Cryo-EM structure of the BG505 SOSIP.664 HIV-1 Env trimer with 3 PGV04 Fabs | | Descriptor: | BG505 SOSIP gp120, BG505 SOSIP gp41, PGV04 heavy chain, ... | | Authors: | Lyumkis, D, Julien, J.-P, Wilson, I.A, Ward, A.B. | | Deposit date: | 2013-10-26 | | Release date: | 2013-11-13 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer.
Science, 342, 2013
|
|
1UTP
 
 | | Trypsin specificity as elucidated by LIE calculations, X-ray structures and association constant measurements | | Descriptor: | 4-PHENYLBUTYLAMINE, CALCIUM ION, GLYCEROL, ... | | Authors: | Leiros, H.-K.S, Brandsdal, B.O, Andersen, O.A, Os, V, Leiros, I, Helland, R, Otlewski, J, Willassen, N.P, Smalas, A.O. | | Deposit date: | 2003-12-09 | | Release date: | 2004-01-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Trypsin Specificity as Elucidated by Lie Calculations, X-Ray Structures, and Association Constant Measurements
Protein Sci., 13, 2004
|
|
1UVQ
 
 | | Crystal structure of HLA-DQ0602 in complex with a hypocretin peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, GLYCINE, ... | | Authors: | Siebold, C, Hansen, B.E, Wyer, J.R, Harlos, K, Esnouf, R.E, Svejgaard, A, Bell, J.I, Strominger, J.L, Jones, E.Y, Fugger, L. | | Deposit date: | 2004-01-22 | | Release date: | 2004-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Hla-Dq0602 that Protects Against Type 1 Diabetes and Confers Strong Susceptibility to Narcolepsy
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2GW3
 
 | | Crystal structure of stony coral fluorescent protein Kaede, green form | | Descriptor: | Kaede, NICKEL (II) ION | | Authors: | Hayashi, I, Mizuno, H, Miyawaki, A, Ikura, M. | | Deposit date: | 2006-05-03 | | Release date: | 2007-05-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic evidence for water-assisted photo-induced peptide cleavage in the stony coral fluorescent protein Kaede.
J.Mol.Biol., 372, 2007
|
|
1V6U
 
 | | Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-alpha-L-arabinofuranosyl-xylobiose | | Descriptor: | alpha-D-xylopyranose, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, beta-D-xylopyranose, ... | | Authors: | Fujimoto, Z, Kaneko, S, Kuno, A, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2003-12-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86
J.Biol.Chem., 279, 2004
|
|
1UN9
 
 | | Crystal structure of the dihydroxyacetone kinase from C. freundii in complex with AMP-PNP and Mg2+ | | Descriptor: | DIHYDROXYACETONE, DIHYDROXYACETONE KINASE, MAGNESIUM ION, ... | | Authors: | Siebold, C, Arnold, I, Garcia-Alles, L.F, Baumann, U, Erni, B. | | Deposit date: | 2003-09-08 | | Release date: | 2003-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the Citrobacter Freundii Dihydroxyacetone Kinase Reveals an Eight-Stranded Alpha-Helical Barrel ATP-Binding Domain
J.Biol.Chem., 278, 2003
|
|
3BB3
 
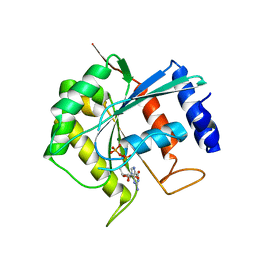 | | Crystal structure of Toc33 from Arabidopsis thaliana in complex with GDP and Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, T7I23.11 protein | | Authors: | Koenig, P, Sinning, I, Schleiff, E, Tews, I. | | Deposit date: | 2007-11-09 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | The GTPase cycle of the chloroplast import receptors Toc33/Toc34: implications from monomeric and dimeric structures.
Structure, 16, 2008
|
|
1V6W
 
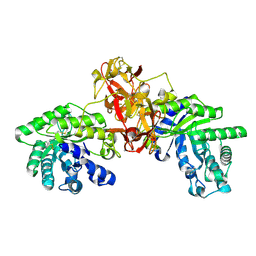 | | Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-4-O-methyl-alpha-D-glucuronosyl-xylobiose | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ENDO-1,4-BETA-D-XYLANASE, beta-D-xylopyranose, ... | | Authors: | Fujimoto, Z, Kaneko, S, Kuno, A, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2003-12-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86
J.Biol.Chem., 279, 2004
|
|
1UXO
 
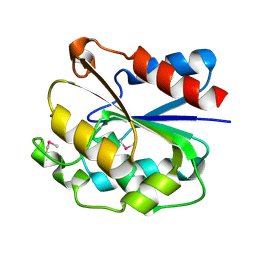 | | The crystal structure of the ydeN gene product from B. subtilis | | Descriptor: | Putative hydrolase YdeN | | Authors: | Janda, I.K, Devedjiev, Y, Cooper, D.R, Chruszcz, M, Derewenda, U, Gabrys, A, Minor, W, Joachimiak, A, Derewenda, Z.S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-27 | | Release date: | 2004-05-27 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Harvesting the high-hanging fruit: the structure of the YdeN gene product from Bacillus subtilis at 1.8 angstroms resolution.
Acta Crystallogr. D Biol. Crystallogr., 60, 2004
|
|
2PMP
 
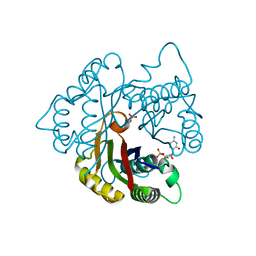 | | Structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from the isoprenoid biosynthetic pathway of Arabidopsis thaliana | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Calisto, B.M, Perez-Gil, J, Querol-Audi, J, Fita, I, Imperial, S. | | Deposit date: | 2007-04-23 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biosynthesis of isoprenoids in plants: Structure of the 2C-methyl-D-erithrytol 2,4-cyclodiphosphate synthase from Arabidopsis thaliana. Comparison with the bacterial enzymes.
Protein Sci., 16, 2007
|
|
