5NKO
 
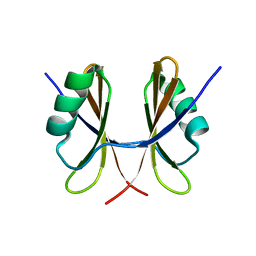 | | Solution structure of the C-terminal domain of S. aureus Hibernating Promoting Factor (CTD-SaHPF) | | Descriptor: | Ribosome hibernation promotion factor | | Authors: | Usachev, K.S, Khusainov, I.S, Ayupov, R.K, Validov, S.Z, Kieffer, B, Yusupov, M.M. | | Deposit date: | 2017-03-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF.
EMBO J., 36, 2017
|
|
8CTI
 
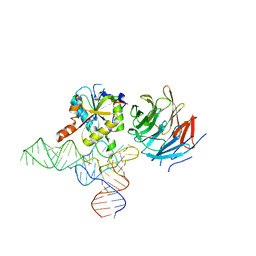 | | Cryo-EM structure of human METTL1-WDR4-tRNA(Val) complex | | Descriptor: | tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4, tRNA-Val-TAC-2-1 | | Authors: | Li, J, Wang, L, Fontana, P, Hunkeler, M, Roy-Burman, S.S, Wu, H, Fischer, E.S, Gregory, R.I. | | Deposit date: | 2022-05-14 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
8CTH
 
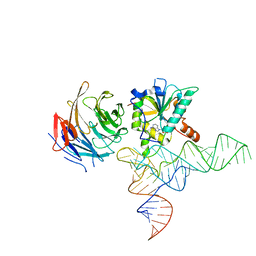 | | Cryo-EM structure of human METTL1-WDR4-tRNA(Phe) complex | | Descriptor: | Phe-tRNA, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(7)-)-methyltransferase, ... | | Authors: | Li, J, Wang, L, Fontana, P, Hunkeler, M, Roy-Burman, S.S, Wu, H, Fishcer, E.S, Gregory, R.I. | | Deposit date: | 2022-05-14 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
4AND
 
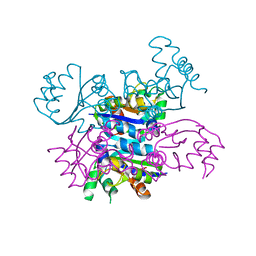 | | CRYSTAL FORM II OF THE D93N MUTANT OF NUCLEOSIDE DIPHOSPHATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Georgescauld, F, Moynie, L, Habersetzer, J, Lascu, I, Dautant, A. | | Deposit date: | 2012-03-16 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Intersubunit Ionic Interactions Stabilize the Nucleoside Diphosphate Kinase of Mycobacterium Tuberculosis.
Plos One, 8, 2013
|
|
6D9K
 
 | | Ternary RsAgo Complex with Guide RNA and Target DNA Containing A-G Non-canonical Pair | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, DNA (5'-D(P*TP*CP*GP*TP*CP*AP*CP*CP*TP*GP*GP*GP*CP*AP*GP*TP*AP*AP*C)-3'), ... | | Authors: | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | Deposit date: | 2018-04-30 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
1J9C
 
 | | Crystal Structure of tissue factor-factor VIIa complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-phenylalaninamide, ... | | Authors: | Huang, M, Ruf, W, Edgington, T.S, Wilson, I.A. | | Deposit date: | 2001-05-24 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand Induced Conformational Transitions of Tissue Factor. Crystal Structure of the Tissue Factor:Factor VIIa Complex.
To be Published
|
|
6DA5
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2R)-1-{[(2S)-1-oxo-3-phenyl-1-{[(pyridin-3-yl)methyl]amino}propan-2-yl]sulfanyl}-3-phenylpropan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Activity Relationships of Rationally Designed Ritonavir Analogues: Impact of Side-Group Stereochemistry, Headgroup Spacing, and Backbone Composition on the Interaction with CYP3A4.
Biochemistry, 58, 2019
|
|
6DAG
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2R)-1-{[(2S)-1-oxo-3-phenyl-1-{[2-(pyridin-3-yl)ethyl]amino}propan-2-yl]sulfanyl}-3-phenylpropan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Activity Relationships of Rationally Designed Ritonavir Analogues: Impact of Side-Group Stereochemistry, Headgroup Spacing, and Backbone Composition on the Interaction with CYP3A4.
Biochemistry, 58, 2019
|
|
8DKT
 
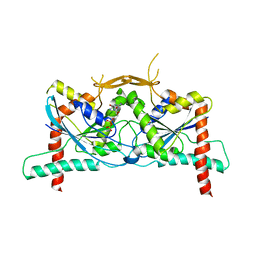 | | Crystal Structure of Septin1 - Septin2 heterocomplex from Drosophila melanogaster | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | de Freitas, A.F, Leonardo, D.A, Cavini, I.A, Pereira, H.M, Garratt, R.C. | | Deposit date: | 2022-07-06 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Conservation and divergence of the G-interfaces of Drosophila melanogaster septins.
Cytoskeleton (Hoboken), 80, 2023
|
|
1LEV
 
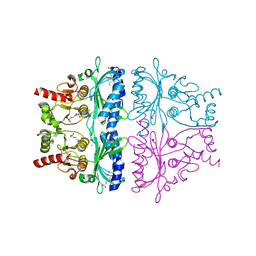 | | PORCINE KIDNEY FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH AN AMP-SITE INHIBITOR | | Descriptor: | 3-(2-CARBOXY-ETHYL)-4,6-DICHLORO-1H-INDOLE-2-CARBOXYLIC ACID, 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, ... | | Authors: | Wright, S.W, Carlo, A.A, Danley, D.E, Hageman, D.L, Karam, G.A, Mansour, M.N, McClure, L.D, Pandit, J, Schulte, G.K, Treadway, J.L, Wang, I.-K, Bauer, P.H. | | Deposit date: | 2002-04-10 | | Release date: | 2002-10-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 3-(2-carboxyethyl)-4,6-dichloro-1H-indole-2-carboxylic acid: an allosteric inhibitor of fructose-1,6-bisphosphatase at the AMP site.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1JE8
 
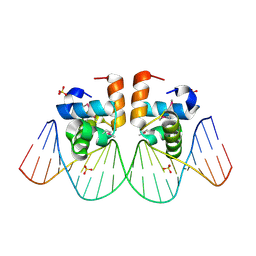 | | Two-Component response regulator NarL/DNA Complex: DNA Bending Found in a High Affinity Site | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*CP*CP*AP*TP*TP*AP*AP*TP*GP*GP*GP*TP*AP*CP*G)-3', Nitrate/Nitrite Response Regulator Protein NARL, SULFATE ION | | Authors: | Maris, A.E, Sawaya, M.R, Kaczor-Grzeskowiak, M, Jarvis, M.R, Bearson, S.M.D, Kopka, M.L, Schroder, I, Gunsalus, R.P, Dickerson, R.E. | | Deposit date: | 2001-06-15 | | Release date: | 2002-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Dimerization allows DNA target site recognition by the NarL response regulator.
Nat.Struct.Biol., 9, 2002
|
|
1JF8
 
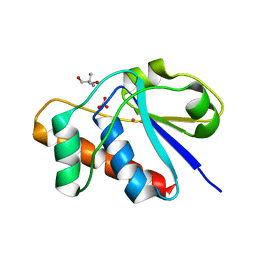 | | X-ray structure of reduced C10S, C15A arsenate reductase from pI258 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICARBONATE ION, POTASSIUM ION, ... | | Authors: | Zegers, I, Martins, J.C, Willem, R, Wyns, L, Messens, J. | | Deposit date: | 2001-06-20 | | Release date: | 2001-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Arsenate reductase from S. aureus plasmid pI258 is a phosphatase drafted for redox duty.
Nat.Struct.Biol., 8, 2001
|
|
8D9N
 
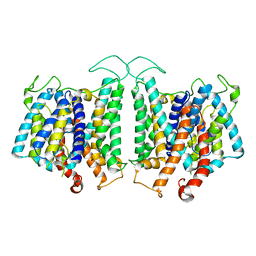 | | CryoEM structures of bAE1 captured in multiple states. | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-06-10 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
1CH4
 
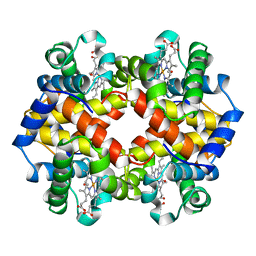 | | MODULE-SUBSTITUTED CHIMERA HEMOGLOBIN BETA-ALPHA (F133V) | | Descriptor: | CARBON MONOXIDE, MODULE-SUBSTITUTED CHIMERA HEMOGLOBIN BETA-ALPHA, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shirai, T, Fujikake, M, Yamane, T, Inaba, K, Ishimori, K, Morishima, I. | | Deposit date: | 1998-06-11 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a protein with an artificial exon-shuffling, module M4-substituted chimera hemoglobin beta alpha, at 2.5 A resolution.
J.Mol.Biol., 287, 1999
|
|
5VTV
 
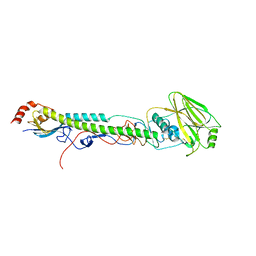 | |
6DM0
 
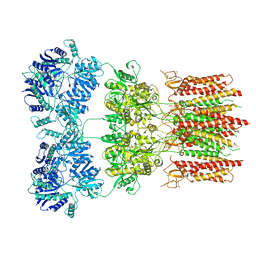 | | Open state GluA2 in complex with STZ and blocked by IEM-1460, after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, ... | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2018-06-04 | | Release date: | 2018-08-22 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
5VTQ
 
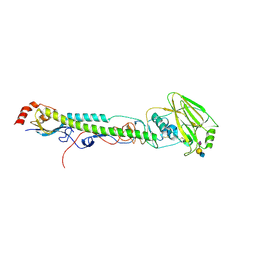 | |
5NWS
 
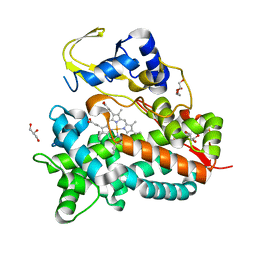 | | Crystal structure of saAcmM involved in actinomycin biosynthesis | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, TETRAETHYLENE GLYCOL, ... | | Authors: | Driller, R, Semsary, S, Crnovicic, I, Vater, J, Keller, U, Loll, B. | | Deposit date: | 2017-05-08 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Ketonization of Proline Residues in the Peptide Chains of Actinomycins by a 4-Oxoproline Synthase.
Chembiochem, 19, 2018
|
|
1JU2
 
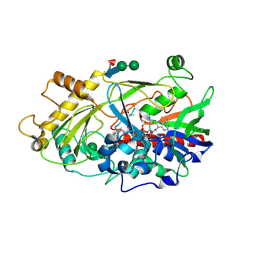 | | Crystal structure of the hydroxynitrile lyase from almond | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dreveny, I, Gruber, K, Glieder, A, Thompson, A, Kratky, C. | | Deposit date: | 2001-08-23 | | Release date: | 2002-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The hydroxynitrile lyase from almond: a lyase that looks like an oxidoreductase.
Structure, 9, 2001
|
|
5W0A
 
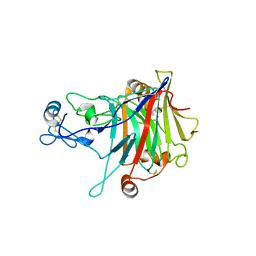 | | Crystal structure of Trichoderma harzianum endoglucanase I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucanase, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Godoy, A.S, Pellegrini, V.O.A, Sonoda, M.T, Kadowaki, M.A, Nascimento, A.S, Polikarpov, I. | | Deposit date: | 2017-05-30 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Structure and dynamics of Trichoderma harzianum Cel7B suggest molecular architecture adaptations required for a wide spectrum of activities on plant cell wall polysaccharides.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
1CQW
 
 | | NAI COCRYSTALLISED WITH HALOALKANE DEHALOGENASE FROM A RHODOCOCCUS SPECIES | | Descriptor: | HALOALKANE DEHALOGENASE; 1-CHLOROHEXANE HALIDOHYDROLASE, IODIDE ION | | Authors: | Newman, J, Peat, T.S, Richard, R, Kan, L, Swanson, P.E, Affholter, J.A, Holmes, I.H, Schindler, J.F, Unkefer, C.J, Terwilliger, T.C. | | Deposit date: | 1999-08-11 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Haloalkane dehalogenases: structure of a Rhodococcus enzyme.
Biochemistry, 38, 1999
|
|
6DA8
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, Nalpha-{(2S)-2-[(tert-butoxycarbonyl)amino]-3-phenylpropyl}-N-[(pyridin-3-yl)methyl]-D-phenylalaninamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure-Activity Relationships of Rationally Designed Ritonavir Analogues: Impact of Side-Group Stereochemistry, Headgroup Spacing, and Backbone Composition on the Interaction with CYP3A4.
Biochemistry, 58, 2019
|
|
3L8Q
 
 | | Structure analysis of the type II cohesin dyad from the adaptor ScaA scaffoldin of Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, Cellulosomal scaffoldin adaptor protein B, ... | | Authors: | Noach, I, Frolow, F, Bayer, E.A. | | Deposit date: | 2010-01-03 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Modular Arrangement of a Cellulosomal Scaffoldin Subunit Revealed from the Crystal Structure of a Cohesin Dyad
J.Mol.Biol., 399, 2010
|
|
5VTX
 
 | |
5VTY
 
 | |
