2A6E
 
 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
7Z6K
 
 | |
2F9L
 
 | | 3D structure of inactive human Rab11b GTPase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAB11B, ... | | Authors: | Scapin, S.M.N, Guimaraes, B.G, Zanchin, N.I.T. | | Deposit date: | 2005-12-06 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of the small GTPase Rab11b reveals critical differences relative to the Rab11a isoform.
J.Struct.Biol., 154, 2006
|
|
5EZM
 
 | | Crystal Structure of ArnT from Cupriavidus metallidurans in the apo state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferases of PMT family, CHLORIDE ION, ... | | Authors: | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-26 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
7Z6A
 
 | |
7Z5Z
 
 | |
7Z5Y
 
 | |
7ZAV
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
6BU1
 
 | |
7ZAW
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
5F91
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator (N-(5-(azepan-1-ylsulfonyl)-2-methoxyphenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Fumarate hydratase class II, ... | | Authors: | Kasbekar, M, Fischer, G, Mott, B.T, Yasgar, A, Hyvonen, M, Boshoff, H.I, Abell, C, Barry, C.E, Thomas, C.J. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Selective small molecule inhibitor of the Mycobacterium tuberculosis fumarate hydratase reveals an allosteric regulatory site.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6MDM
 
 | | The 20S supercomplex engaging the SNAP-25 N-terminus (class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Alpha-soluble NSF attachment protein, ... | | Authors: | White, K.I, Zhao, M, Brunger, A.T. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural principles of SNARE complex recognition by the AAA+ protein NSF.
Elife, 7, 2018
|
|
4CAG
 
 | | Bacillus licheniformis Rhamnogalacturonan Lyase PL11 | | Descriptor: | CALCIUM ION, GLYCEROL, POLYSACCHARIDE LYASE FAMILY 11 PROTEIN | | Authors: | Otten, H, Rodrigues da Silva, I.I.C, Jers, C, Nyffenegger, C, Larsen, D.M, Mikkelsen, J.D, Larsen, S. | | Deposit date: | 2013-10-08 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Design of Thermostable Rhamnogalacturonan Lyase Mutants from Bacillus Licheniformis by Combination of Targeted Single Point Mutations.
Appl.Microbiol.Biotechnol., 98, 2014
|
|
6AME
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 M21A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1T4K
 
 | | Crystal Structure of Unliganded Aldolase Antibody 93F3 Fab | | Descriptor: | IMMUNOGLOBULIN IGG1, HEAVY CHAIN, KAPPA LIGHT CHAIN, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2004-04-29 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Origin of Enantioselectivity in Aldolase Antibodies: Crystal Structure, Site-directed Mutagenesis, and Computational Analysis
J.Mol.Biol., 343, 2004
|
|
5IV8
 
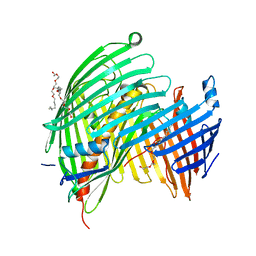 | | The LPS Transporter LptDE from Klebsiella pneumoniae, core complex | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LPS biosynthesis protein, LPS-assembly lipoprotein LptE | | Authors: | Botos, I, McCarthy, J.G, Buchanan, S.K. | | Deposit date: | 2016-03-20 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.938 Å) | | Cite: | Structural and Functional Characterization of the LPS Transporter LptDE from Gram-Negative Pathogens.
Structure, 24, 2016
|
|
7Z04
 
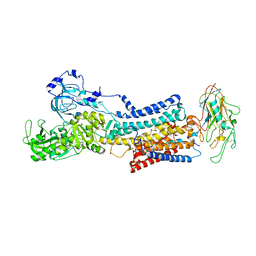 | | 10 mM Rb+ soak of beryllium fluoride inhibited Na+,K+-ATPase, E2-BeFx (rigid body model) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, FXYD domain-containing ion transport regulator, MAGNESIUM ION, ... | | Authors: | Fruergaard, M.U, Dach, I, Andersen, J.L, Ozol, M, Shasavar, A, Quistgaard, E.M, Poulsen, H, Fedosova, N.U, Nissen, P. | | Deposit date: | 2022-02-22 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (7.5 Å) | | Cite: | The Na + ,K + -ATPase in complex with beryllium fluoride mimics an ATPase phosphorylated state.
J.Biol.Chem., 298, 2022
|
|
7YZR
 
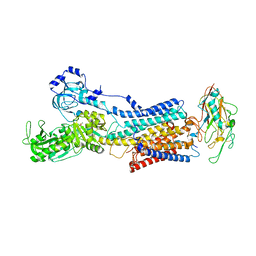 | | 50 mM Rb+ soak of beryllium fluoride inhibited Na+,K+-ATPase, E2-BeFx (rigid body model) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, FXYD domain-containing ion transport regulator, MAGNESIUM ION, ... | | Authors: | Fruergaard, M.U, Dach, I, Andersen, J.L, Ozol, M, Shasavar, A, Quistgaard, E.M, Poulsen, H, Fedosova, N.U, Nissen, P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (6.92 Å) | | Cite: | The Na + ,K + -ATPase in complex with beryllium fluoride mimics an ATPase phosphorylated state.
J.Biol.Chem., 298, 2022
|
|
2JX9
 
 | |
7EC7
 
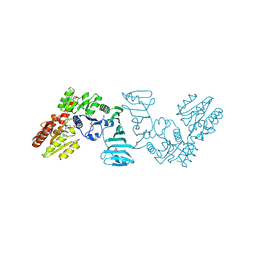 | | Crystal structure of SdgB (complexed with phosphate ions) | | Descriptor: | Glycosyl transferase, group 1 family protein, PHOSPHATE ION | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5J5Q
 
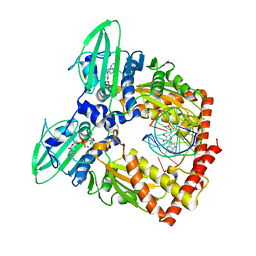 | | AMP-PNP-stabilized ATPase domain of topoisomerase IV from Streptococcus pneumoniae, complex type II | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*AP*TP*AP*TP*AP*TP*AP*TP*GP*C)-3'), DNA topoisomerase 4 subunit B, MAGNESIUM ION, ... | | Authors: | Laponogov, I, Pan, X.-S, Skamrova, G, Umrekar, T, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2016-04-03 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Trapping of the transport-segment DNA by the ATPase domains of a type II topoisomerase.
Nat Commun, 9, 2018
|
|
3IHY
 
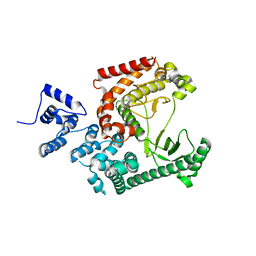 | | Human PIK3C3 crystal structure | | Descriptor: | Phosphatidylinositol 3-kinase catalytic subunit type 3 | | Authors: | Siponen, M.I, Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Kotzsch, A, Kragh Nielsen, T, Moche, M, Nyman, T, Persson, C, Roos, A.K, Sagemark, J, Schueler, H, Schutz, P, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-31 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human PIK3C3 crystal structure
TO BE PUBLISHED
|
|
4IV3
 
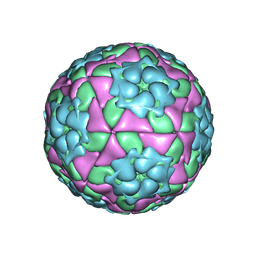 | | Crystal structure of recombinant foot-and-mouth-disease virus A22-H2093C empty capsid | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Porta, C, Kotecha, A, Burman, A, Jackson, T, Ren, J, Loureiro, S, Jones, I.M, Fry, E.E, Stuart, D.I, Charleston, B. | | Deposit date: | 2013-01-22 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rational engineering of recombinant picornavirus capsids to produce safe, protective vaccine antigen.
Plos Pathog., 9, 2013
|
|
7EC3
 
 | | Crystal structure of SdgB (complexed with UDP, GlcNAc, and Glycosylated peptide) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-35)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-65)]5,6-DIHYDRO-BENZO[H]CINNOLIN-3-YLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyl transferase, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3IPY
 
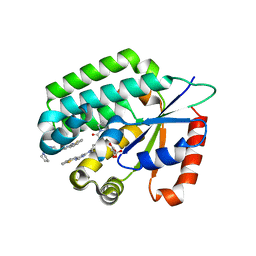 | | X-Ray structure of Human Deoxycytidine Kinase in complex with an inhibitor | | Descriptor: | 4-(1-benzothiophen-2-yl)-6-[4-(2-oxo-2-pyrrolidin-1-ylethyl)piperazin-1-yl]pyrimidine, D-MALATE, Deoxycytidine kinase | | Authors: | Tari, L.W, Swanson, R.V, Hunter, M, Hoffman, I, Stouch, T.R, Carson, K.G. | | Deposit date: | 2009-08-18 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Lead optimization and structure-based design of potent and bioavailable deoxycytidine kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
