8P6Y
 
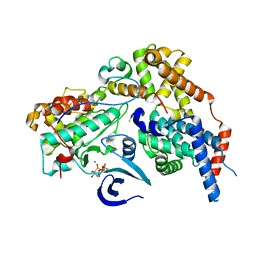 | | Cryo-EM structure of CAK in complex with nucleotide analogue ATPgS | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
6RYD
 
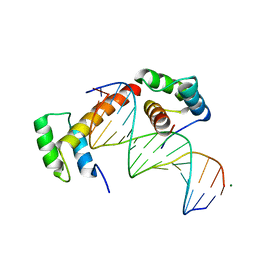 | | WUS-HD bound to TGAA DNA | | Descriptor: | DNA (5'-D(*AP*GP*TP*GP*TP*AP*TP*GP*AP*AP*TP*GP*AP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*TP*CP*AP*TP*TP*CP*AP*TP*AP*CP*AP*CP*T)-3'), MAGNESIUM ION, ... | | Authors: | Sloan, J.J, Wild, K, Sinning, I. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.575 Å) | | Cite: | Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL.
Nat Commun, 11, 2020
|
|
4RJ2
 
 | |
2GQK
 
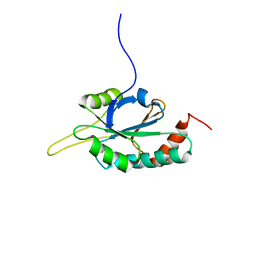 | | Solution structure of Human Ni(II)-Sco1 | | Descriptor: | NICKEL (II) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-21 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1TG7
 
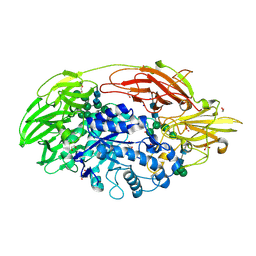 | | Native structure of beta-galactosidase from Penicillium sp. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rojas, A.L, Nagem, R.A.P, Neustroev, K.N, Arand, M, Adamska, M, Eneyskaya, E.V, Kulminskaya, A.A, Garratt, R.C, Golubev, A.M, Polikarpov, I. | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of beta-Galactosidase from Penicillium sp. and its Complex with Galactose
J.Mol.Biol., 343, 2004
|
|
2V4A
 
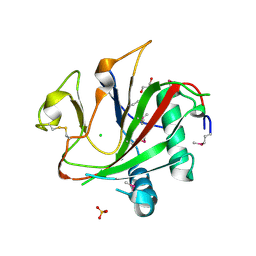 | | Crystal structure of the SeMet-labeled prolyl-4 hydroxylase (P4H) type I from green algae Chlamydomonas reinhardtii. | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koski, M.K, Hieta, R, Bollner, C, Kivirikko, K.I, Myllyharju, J, Wierenga, R.K. | | Deposit date: | 2007-06-28 | | Release date: | 2007-10-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Active Site of an Algal Prolyl 4-Hydroxylase Has a Large Structural Plasticity.
J.Biol.Chem., 282, 2007
|
|
6RRE
 
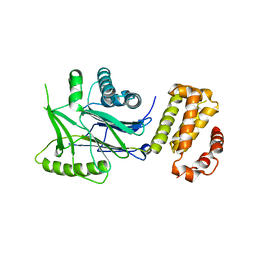 | | SidD, deAMPylase from Legionella pneumophila | | Descriptor: | Adenosine monophosphate-protein hydrolase SidD, MAGNESIUM ION | | Authors: | Tascon, I, Lucas, M, Rojas, A.L, Hierro, A. | | Deposit date: | 2019-05-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.586 Å) | | Cite: | Structural insight into the membrane targeting domain of the Legionella deAMPylase SidD.
Plos Pathog., 16, 2020
|
|
6RTB
 
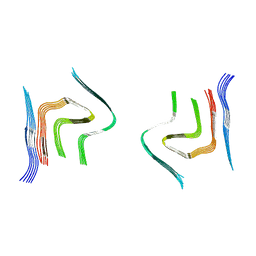 | | cryo-em structure of alpha-synuclein fibril polymorph 2B | | Descriptor: | Alpha-synuclein | | Authors: | Guerrero-Ferreira, R, Taylor, N.M.I, Arteni, A.A, Melki, R, Meier, B.H, Bockmann, A, Bousset, L, Stahlberg, H. | | Deposit date: | 2019-05-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy.
Elife, 8, 2019
|
|
6RUX
 
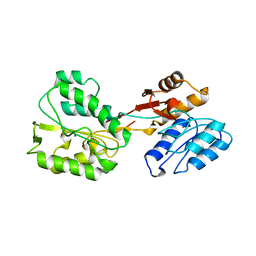 | | P46, an immunodominant surface protein from Mycoplasma hyopneumoniae | | Descriptor: | 46 kDa surface antigen, SODIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Guasch, A, Gonzalez-Gonzalez, L, Fita, I. | | Deposit date: | 2019-05-29 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of P46, an immunodominant surface protein from Mycoplasma hyopneumoniae: interaction with a monoclonal antibody.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
2GVN
 
 | | L-asparaginase from Erwinia carotovora in complex with aspartic acid | | Descriptor: | ASPARTIC ACID, L-asparaginase | | Authors: | Kravchenko, O.V, Kislitsin, Y.A, Popov, A.N, Nikonov, S.V, Kuranova, I.P. | | Deposit date: | 2006-05-03 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-dimensional structures of L-asparaginase from Erwinia carotovora complexed with aspartate and glutamate.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
8P6V
 
 | | Cryo-EM structure of CAK in complex with inhibitor ICEC0942 | | Descriptor: | (3R,4R)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8OGG
 
 | | Crystal structure of FutA after an accumulated dose of 5 kGy | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3GMQ
 
 | | Structure of mouse CD1d expressed in SF9 cells, no ligand added | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2 microglobulin, ... | | Authors: | Schiefner, A, Wilson, I.A. | | Deposit date: | 2009-03-14 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
8ORM
 
 | | Cryo-EM structure of CAK-THZ1 | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-04-14 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
6S74
 
 | | Crystal structure of CARM1 in complex with inhibitor UM305 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-azanylpropyl-[3-(pyrimidin-2-ylamino)propyl]amino]methyl]oxolane-3,4-diol, GLYCEROL, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
6CI6
 
 | | Crystal structure of equine serum albumin in complex with nabumetone | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, Serum albumin, ... | | Authors: | Venkataramany, B.S, Czub, M.P, Shabalin, I.G, Handing, K.B, Steen, E.H, Cooper, D.R, Joachimiak, A, Satchell, K.J.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Albumin-Based Transport of Nonsteroidal Anti-Inflammatory Drugs in Mammalian Blood Plasma.
J.Med.Chem., 2020
|
|
3N8E
 
 | | Substrate binding domain of the human Heat Shock 70kDa protein 9 (mortalin) | | Descriptor: | Stress-70 protein, mitochondrial | | Authors: | Wisniewska, M, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, van der Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-28 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate binding domain of the human Heat Shock 70kDa protein 9 (mortalin)
To be published
|
|
6UYM
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2mc3-v6 redesigned core from genotype 1a bound to broadly neutralizing antibody AR3C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Zhu, J. | | Deposit date: | 2019-11-13 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.848 Å) | | Cite: | Proof of concept for rational design of hepatitis C virus E2 core nanoparticle vaccines.
Sci Adv, 6, 2020
|
|
6CK8
 
 | |
1QJU
 
 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND VP61209 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[2N-METHYL-2H-TETRAZOL-5-YL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Minor, I, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-05 | | Release date: | 1999-07-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6RKY
 
 | |
7SKR
 
 | | BtSCoV-Rf1.2004 Papain-Like protease bound to the non-covalent inhibitor 37 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(2-methoxypyridin-4-yl)methyl]-2-[(1R)-1-(naphthalen-1-yl)ethyl]-2-azaspiro[3.3]heptane-6-carboxamide, ... | | Authors: | Durie, I, Shepard, J, Freitas, B, O'Boyle, B, Enos, S, Pegan, S.D. | | Deposit date: | 2021-10-21 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Exploring Noncovalent Protease Inhibitors for the Treatment of Severe Acute Respiratory Syndrome and Severe Acute Respiratory Syndrome-Like Coronaviruses.
Acs Infect Dis., 8, 2022
|
|
1UPM
 
 | | ACTIVATED SPINACH RUBISCO COMPLEXED WITH 2-CARBOXYARABINITOL 2 BISPHOSPHAT AND CA2+. | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, CALCIUM ION, RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN, ... | | Authors: | Karkehabadi, S, Taylor, T.C, Andersson, I. | | Deposit date: | 2003-10-08 | | Release date: | 2003-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Calcium Supports Loop Closure But not Catalysis in Rubisco
J.Mol.Biol., 334, 2003
|
|
5NOZ
 
 | | Structure of cyclophilin A in complex with 3,4-diaminobenzohydrazide | | Descriptor: | 3,4-bis(azanyl)benzohydrazide, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
6RO6
 
 | | Crystal structure of the C-terminal dimerization domain of the essential repressor DdrO from radiation-resistant Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | HTH-type transcriptional regulator DdrOC, SULFATE ION | | Authors: | Pignol, D, Arnoux, P, Siponen, M.I, Brandelet, G, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
