5KCX
 
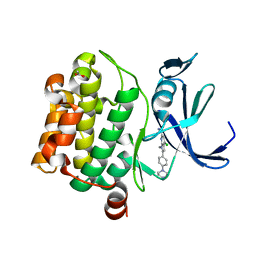 | | Pim-1 kinase in Complex with a Selective N-substituted 7-azaindole Inhibitor | | Descriptor: | 4-chloranyl-1-methyl-2-[4-(4-methylpiperazin-1-yl)phenyl]pyrrolo[2,3-b]pyridine-6-carboxamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Mechin, I, McLean, L.R, Zhang, Y, Wang, R. | | Deposit date: | 2016-06-07 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of N-substituted 7-azaindoles as PIM1 kinase inhibitors - Part I.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
3RU2
 
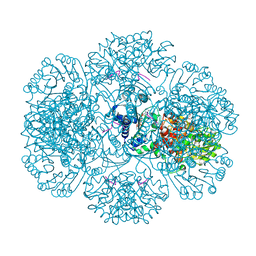 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADPH. | | Descriptor: | BETA-6-HYDROXY-1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-04 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RXA
 
 | | Crystal structure of Trypsin complexed with cycloheptanamine | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXK
 
 | | Crystal structure of Trypsin complexed with methyl 4-amino-1-methyl-pyrrolidine-2-carboxylate | | Descriptor: | CALCIUM ION, Cationic trypsin, DIMETHYL SULFOXIDE, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
4LDQ
 
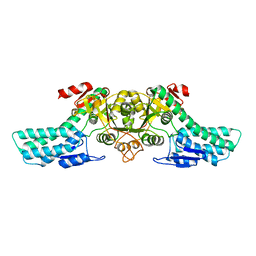 | |
5JQ7
 
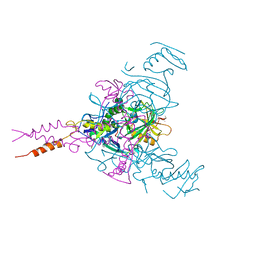 | | Crystal structure of Ebola glycoprotein in complex with toremifene | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Envelope glycoprotein 1,Envelope glycoprotein 1,Envelope glycoprotein 1, ... | | Authors: | Zhao, Y, Ren, J, Stuart, D.I. | | Deposit date: | 2016-05-04 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Toremifene interacts with and destabilizes the Ebola virus glycoprotein.
Nature, 535, 2016
|
|
4LDO
 
 | | Structure of beta2 adrenoceptor bound to adrenaline and an engineered nanobody | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, Camelid Antibody Fragment, L-EPINEPHRINE, ... | | Authors: | Ring, A.M, Manglik, A, Kruse, A.C, Enos, M.D, Weis, W.I, Garcia, K.C, Kobilka, B.K. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody.
Nature, 502, 2013
|
|
5KCE
 
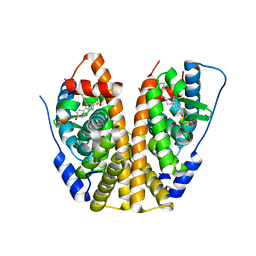 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-methyl, 2-chlorobenzyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-N-(2-chlorophenyl)-5,6-bis(4-hydroxyphenyl)-N-methyl-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
1WQ2
 
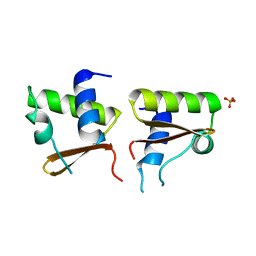 | | Neutron Crystal Structure Of Dissimilatory Sulfite Reductase D (DsrD) | | Descriptor: | Protein dsvD, SULFATE ION | | Authors: | Chatake, T, Mizuno, N, Voordouw, G, Higuchi, Y, Arai, S, Tanaka, I, Niimura, N. | | Deposit date: | 2004-09-19 | | Release date: | 2005-09-19 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2.4 Å) | | Cite: | Crystallization and preliminary neutron analysis of the dissimilatory sulfite reductase D (DsrD) protein from the sulfate-reducing bacterium Desulfovibrio vulgaris.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4LJ8
 
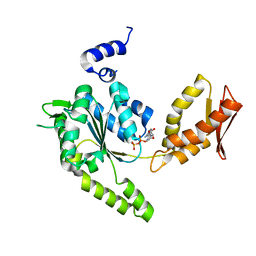 | | ClpB NBD2 R621Q from T. thermophilus in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4L9J
 
 | |
4L9T
 
 | |
4LAS
 
 | | Crystal structure of a therapeutic single chain antibody in complex with 4-hydroxymethamphetamine | | Descriptor: | 4-[(2S)-2-(methylamino)propyl]phenol, Single chain antibody fragment scFv6H4 | | Authors: | Celical, R, Gokulan, K, Peterson, E.C, Varughese, K.I. | | Deposit date: | 2013-06-20 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural characterization of a therapeutic anti-methamphetamine antibody fragment: oligomerization and binding of active metabolites.
Plos One, 8, 2013
|
|
4LNW
 
 | | Crystal structure of TR-alpha bound to T3 in a second site | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Thyroid hormone receptor alpha | | Authors: | Puhl, A.C, Aparicio, R, Polikarpov, I. | | Deposit date: | 2013-07-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a new hormone-binding site on the surface of thyroid hormone receptor.
Mol.Endocrinol., 28, 2014
|
|
4LDE
 
 | | Structure of beta2 adrenoceptor bound to BI167107 and an engineered nanobody | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid Antibody Fragment, ... | | Authors: | Ring, A.M, Manglik, A, Kruse, A.C, Enos, M.D, Weis, W.I, Garcia, K.C, Kobilka, B.K. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody.
Nature, 502, 2013
|
|
6IOQ
 
 | | The ligand binding domain of Mlp24 with glycine | | Descriptor: | CALCIUM ION, GLYCINE, Methyl-accepting chemotaxis protein | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
3SJB
 
 | | Crystal structure of S. cerevisiae Get3 in the open state in complex with Get1 cytosolic domain | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 1, PHOSPHATE ION, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
3RXR
 
 | | Crystal structure of Trypsin complexed with cycloheptanamine (F01 and F03, cocktail experiment) | | Descriptor: | CALCIUM ION, Cationic trypsin, DIMETHYL SULFOXIDE, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
9FBD
 
 | | Crystal structure of 3-hydroxybutyryl-CoA dehydrogenase from Thermus thermophilus HB27 complexed to NAD+ | | Descriptor: | 3-hydroxybutyryl-CoA dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hurtado-Guerrero, R, Macias-Leon, J, Gines-Alcober, I, Gonzalez-Ramirez, A.M. | | Deposit date: | 2024-05-13 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Loop engineering of enzymes to control their immobilization and ultimately fabricate more efficient heterogeneous biocatalysts.
Protein Sci., 34, 2025
|
|
5KCU
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, alpha-naphthyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(naphthalen-2-yl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KEL
 
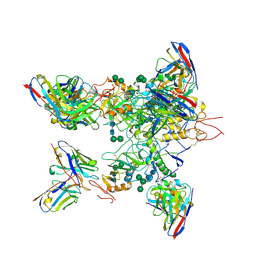 | | EBOV GP in complex with variable Fab domains of IgGs c2G4 and c13C6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ebola surface glycoprotein, ... | | Authors: | Pallesen, J, Murin, C.D, de Val, N, Cottrell, C.A, Hastie, K.M, Turner, H.L, Fusco, M.L, Flyak, A.I, Zeitlin, L, Crowe Jr, J.E, Andersen, K.G, Saphire, E.O, Ward, A.B. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of Ebola virus GP and sGP in complex with therapeutic antibodies.
Nat Microbiol, 1, 2016
|
|
9F38
 
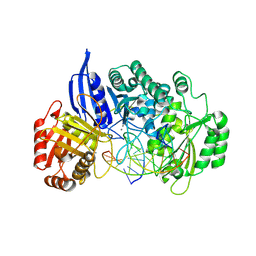 | | BsmI (wild-type) crystallized with Ca2+ and cognate dsDNA | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BsmI, CALCIUM ION, ... | | Authors: | Sieskind, R, Missoury, S, Madru, C, Commenge, I, Niogret, G, Hollenstein, M, Rondelez, Y, Haouz, A, Sauguet, L, Legrand, P, Delarue, M. | | Deposit date: | 2024-04-25 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structures of monomeric BsmI restriction endonuclease reveal coordinated sequential cleavage of two DNA strands.
Commun Biol, 8, 2025
|
|
3S5Y
 
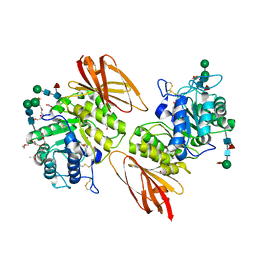 | | Pharmacological Chaperoning in Human alpha-Galactosidase | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guce, A.I, Clark, N.E, Garman, S.C. | | Deposit date: | 2011-05-23 | | Release date: | 2012-01-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | The molecular basis of pharmacological chaperoning in human alpha-galactosidase
Chem.Biol., 18, 2011
|
|
9EZ7
 
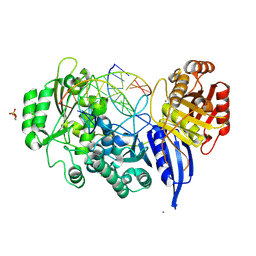 | | BsmI (Nicking top mutant) crystallized with Ca2+ and cognate dsDNA | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BsmI, CALCIUM ION, ... | | Authors: | Sieskind, R, Missoury, S, Madru, C, Commenge, I, Niogret, G, Hollenstein, M, Rondelez, Y, Haouz, A, Legrand, P, Sauguet, L, Delarue, M. | | Deposit date: | 2024-04-10 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Crystal structures of monomeric BsmI restriction endonuclease reveal coordinated sequential cleavage of two DNA strands.
Commun Biol, 8, 2025
|
|
9EZD
 
 | | BsmI (Bottom Nicking mutant) crystallized with Mg2+ and cognate dsDNA (Post-reactive complex) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BsmI, CHLORIDE ION, ... | | Authors: | Sieskind, R, Missoury, S, Madru, C, Commenge, I, Niogret, G, Rondelez, Y, Haouz, A, Legrand, P, Sauguet, L, Delarue, M. | | Deposit date: | 2024-04-11 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.635 Å) | | Cite: | Crystal structures of monomeric BsmI restriction endonuclease reveal coordinated sequential cleavage of two DNA strands.
Commun Biol, 8, 2025
|
|
