1DGP
 
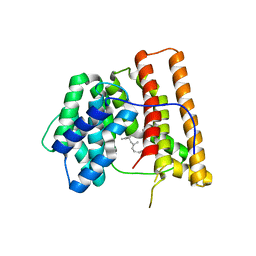 | | ARISTOLOCHENE SYNTHASE FARNESOL COMPLEX | | Descriptor: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, ARISTOLOCHENE SYNTHASE | | Authors: | Caruthers, J.M, Kang, I, Cane, D.E, Christianson, D.W. | | Deposit date: | 1999-11-24 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure determination of aristolochene synthase from the blue cheese mold, Penicillium roqueforti.
J.Biol.Chem., 275, 2000
|
|
2L0W
 
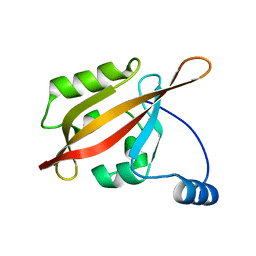 | | Solution NMR structure of the N-terminal PAS domain of HERG potassium channel | | Descriptor: | Potassium voltage-gated channel, subfamily H (Eag-related), member 2, ... | | Authors: | Ng, C.A, Hunter, M.J, Mobli, M, King, G.F, Kuchel, P.W, Vandenberg, J.I. | | Deposit date: | 2010-07-19 | | Release date: | 2011-01-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The N-Terminal Tail of hERG Contains an Amphipathic alpha-Helix That Regulates Channel Deactivation
PLoS ONE, 6, 2011
|
|
2L8J
 
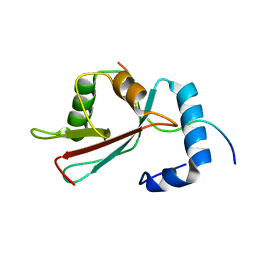 | | GABARAPL-1 NBR1-LIR complex structure | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, NBR1-LIR peptide | | Authors: | Rogov, V.V, Rozenknop, A, Rogova, N.Y, Loehr, F, Guentert, P, Dikic, I, Doetsch, V. | | Deposit date: | 2011-01-17 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of the Interaction of GABARAPL-1 with the LIR Motif of NBR1.
J.Mol.Biol., 410, 2011
|
|
2LD7
 
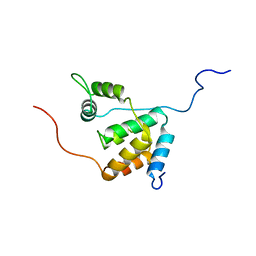 | | Solution structure of the mSin3A PAH3-SAP30 SID complex | | Descriptor: | Histone deacetylase complex subunit SAP30, Paired amphipathic helix protein Sin3a | | Authors: | Xie, T, He, Y, Korkeamaki, H, Zhang, Y, Imhoff, R, Lohi, O, Radhakrishnan, I. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the 30-kDa Sin3-associated protein (SAP30) in complex with the mammalian Sin3A corepressor and its role in nucleic acid binding.
J.Biol.Chem., 286, 2011
|
|
3SZS
 
 | |
5N9X
 
 | | Structure of adenylation domain THR1 involved in the biosynthesis of 4-chlorothreonine in Streptomyces SP.OH-5093, ligand bound structure | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenylation domain, MAGNESIUM ION, ... | | Authors: | Savino, C, Vallone, B, Scaglione, A, Parisi, G, Montemiglio, L.C, Fullone, M.R, Grgurina, I. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structure of the adenylation domain Thr1 involved in the biosynthesis of 4-chlorothreonine in Streptomyces sp. OH-5093-protein flexibility and molecular bases of substrate specificity.
FEBS J., 284, 2017
|
|
2LQG
 
 | | Solution Structure of the R4 domain of talin | | Descriptor: | Talin-1 | | Authors: | Goult, B.T, Gingras, A.R, Bate, N, Roberts, G.C.K, Barsukov, I.L, Critchley, D.R. | | Deposit date: | 2012-03-06 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RIAM and Vinculin Binding to Talin Are Mutually Exclusive and Regulate Adhesion Assembly and Turnover.
J.Biol.Chem., 288, 2013
|
|
1DF8
 
 | | S45A MUTANT OF STREPTAVIDIN IN COMPLEX WITH BIOTIN | | Descriptor: | BIOTIN, PROTEIN (STREPTAVIDIN) | | Authors: | Hyre, D.E, Le Trong, I, Freitag, S, Stenkamp, R.E, Stayton, P.S. | | Deposit date: | 1999-11-18 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Ser45 plays an important role in managing both the equilibrium and transition state energetics of the streptavidin-biotin system.
Protein Sci., 9, 2000
|
|
2LSY
 
 | |
2L85
 
 | | Solution NMR structures of CBP bromodomain with small molecule of HBS | | Descriptor: | 4-[(E)-(4-hydroxyphenyl)diazenyl]benzenesulfonic acid, CREB-binding protein | | Authors: | Borah, J.C, Mujtaba, S, Karakikes, I, Zeng, L, Muller, M, Patel, J, Moshkina, N, Morohashi, K, Zhang, W, Gerona-Navarro, G, Hajjar, R.J, Zhou, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule Binding to the Coactivator CREB-Binding Protein Blocks Apoptosis in Cardiomyocytes.
Chem.Biol., 18, 2011
|
|
3SRK
 
 | | A new class of suicide inhibitor blocks nucleotide binding to pyruvate kinase | | Descriptor: | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE, POTASSIUM ION, Pyruvate kinase, ... | | Authors: | Morgan, H.P, Walsh, M, Blackburn, E.A, Wear, M.A, Boxer, M, Shen, M, McNae, I.W, Michels, P.A.M, Auld, D.S, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-07-07 | | Release date: | 2012-07-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A new class of suicide inhibitor blocks nucleotide binding to pyruvate kinase
To be Published
|
|
5NP3
 
 | | Abl2 SH3 | | Descriptor: | Abelson tyrosine-protein kinase 2 | | Authors: | Mero, B, Radnai, L, Gogl, G, Leveles, I, Buday, L. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the tyrosine phosphorylation-mediated inhibition of SH3 domain-ligand interactions.
J.Biol.Chem., 294, 2019
|
|
2LM0
 
 | | Solution structure of the AF4-AF9 complex | | Descriptor: | AF4/FMR2 family member 1/Protein AF-9 chimera | | Authors: | Leach, B.I, Kuntimaddi, A, Schmidt, C.R, Cierpicki, T, Johnson, S.A, Bushweller, J.H. | | Deposit date: | 2011-11-18 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Leukemia Fusion Target AF9 Is an Intrinsically Disordered Transcriptional Regulator that Recruits Multiple Partners via Coupled Folding and Binding.
Structure, 21, 2013
|
|
1EEA
 
 | | Acetylcholinesterase | | Descriptor: | PROTEIN (ACETYLCHOLINESTERASE) | | Authors: | Raves, M.L, Giles, K, Schrag, J.D, Schmid, M.F, Phillips Jr, G.N, Wah, C, Howard, A.J, Silman, I, Sussman, J.L. | | Deposit date: | 1999-01-26 | | Release date: | 1999-02-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Quaternary Structure of Tetrameric Acetylcholinesterase
Structure and Function of Cholinesterases and Related Proteins, 1998
|
|
2L50
 
 | | Solution structure of apo S100A16 | | Descriptor: | Protein S100-A16 | | Authors: | Babini, E, Bertini, I, Borsi, V, Calderone, V, Hu, X, Luchinat, C, Parigi, G. | | Deposit date: | 2010-10-22 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of human S100A16, a low-affinity calcium binder.
J.Biol.Inorg.Chem., 16, 2011
|
|
2LLS
 
 | | solution structure of human apo-S100A1 C85M | | Descriptor: | Protein S100-A1 | | Authors: | Budzinska, M, Jaremko, L, Jaremko, M, Zdanowski, K, Zhukov, I, Bierzynski, A, Ejchart, A. | | Deposit date: | 2011-11-17 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Chemical Shift Assignments and solution structure of human apo-S100A1 C85M mutant
To be Published
|
|
1EJU
 
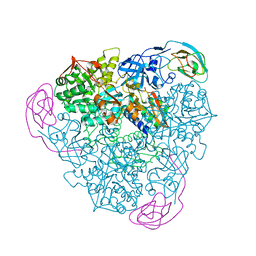 | | CRYSTAL STRUCTURE OF THE H320N VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | NICKEL (II) ION, UREASE ALPHA SUBUNIT, UREASE BETA SUBUNIT, ... | | Authors: | Pearson, M.A, Park, I.S, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 2000-03-04 | | Release date: | 2000-09-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural characterization of urease active site variants.
Biochemistry, 39, 2000
|
|
5N71
 
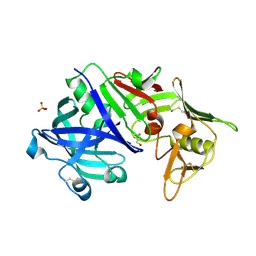 | | CRYSTAL STRUCTURE OF MATURE CATHEPSIN D FROM THE TICK IXODES RICINUS (IRCD1) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative cathepsin d, SULFATE ION | | Authors: | Brynda, J, Hanova, I, Hobizalova, R, Mares, M. | | Deposit date: | 2017-02-17 | | Release date: | 2017-12-27 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Novel Structural Mechanism of Allosteric Regulation of Aspartic Peptidases via an Evolutionarily Conserved Exosite.
Cell Chem Biol, 25, 2018
|
|
2LRJ
 
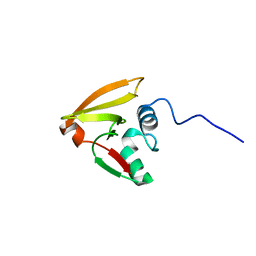 | |
4XQU
 
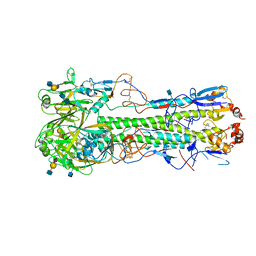 | | Crystal structure of hemagglutinin from Jiangxi-Donghu (2013) H10N8 influenza virus in complex with 3'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Tzarum, N, Zhang, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2015-01-20 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | A Human-Infecting H10N8 Influenza Virus Retains a Strong Preference for Avian-type Receptors.
Cell Host Microbe, 17, 2015
|
|
5NE0
 
 | | Room temperature in-situ structure of hen egg-white lysozyme from crystals enclosed between ultrathin silicon nitride membranes | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Martiel, I, Opara, N, Arnold, S.A, Braun, T, Stahlberg, H, Makita, M, David, C, Padeste, C. | | Deposit date: | 2017-03-09 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Direct protein crystallization on ultrathin membranes for diffraction measurements at X-ray free-electron lasers.
J.Appl.Crystallogr., 50, 2017
|
|
1E99
 
 | | Human thymidylate kinase complexed with AZTMP and ADP | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostermann, N, Lavie, A, Padiyar, S, Brundiers, R, Veit, T, Reinstein, J, Goody, R.S, Konrad, M, Schlichting, I. | | Deposit date: | 2000-10-10 | | Release date: | 2001-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potentiating Azt Activation: Structures of Wildtype and Mutant Human Thymidylate Kinase Suggest Reasons for the Mutants' Improved Kinetics with the HIV Prodrug Metabolite Aztmp
J.Mol.Biol., 304, 2000
|
|
2L8D
 
 | | Structure/function of the LBR Tudor domain | | Descriptor: | Lamin-B receptor | | Authors: | Liokatis, S, Edlich, C, Soupsana, K, Giannios, I, Sattler, M, Georgatos, S.D, Politou, A.S. | | Deposit date: | 2011-01-10 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and molecular interactions of lamin B receptor tudor domain.
J.Biol.Chem., 287, 2012
|
|
2LOQ
 
 | | Backbone structure of human membrane protein FAM14B (Interferon alpha-inducible protein 27-like protein 1) | | Descriptor: | Interferon alpha-inducible protein 27-like protein 1 | | Authors: | Klammt, C, Chui, E.J, Maslennikov, I, Kwiatkowski, W, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
5NPI
 
 | |
