1XW5
 
 | |
6M3N
 
 | | Solution structure of anti-CRISPR AcrIF7 | | Descriptor: | anti-CRIPSR AcrIF7 | | Authors: | Kim, I, An, S.Y, Koo, J, Bae, E, Suh, J.Y. | | Deposit date: | 2020-03-04 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and mechanistic insights into the CRISPR inhibition of AcrIF7.
Nucleic Acids Res., 48, 2020
|
|
3FHP
 
 | | A neutron crystallographic analysis of a porcine 2Zn insulin at 2.0 A resolution | | Descriptor: | Insulin, ZINC ION | | Authors: | Iwai, W, Kurihara, K, Yamada, T, Kobayashi, Y, Ohnishi, Y, Tanaka, I, Takahashi, H, Niimura, N. | | Deposit date: | 2008-12-09 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | A neutron crystallographic analysis of T6 porcine insulin at 2.1 A resolution
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1UZK
 
 | | Integrin binding cbEGF22-TB4-cbEGF33 fragment of human fibrillin-1, Ca bound to cbEGF23 domain only | | Descriptor: | CALCIUM ION, FIBRILLIN-1 | | Authors: | Lee, S.S.J, Knott, V, Harlos, K, Handford, P.A, Stuart, D.I. | | Deposit date: | 2004-03-13 | | Release date: | 2006-05-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the Integrin Binding Fragment from Fibrillin-1 Gives New Insights Into Microfibril Organization
Structure, 12, 2004
|
|
4HS7
 
 | | 2.6 Angstrom Structure of the Extracellular Solute-binding Protein from Staphylococcus aureus in complex with PEG. | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Bacterial extracellular solute-binding protein, putative, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 Angstrom Structure of the Extracellular Solute-binding Protein from Staphylococcus aureus in complex with PEG.
TO BE PUBLISHED
|
|
1TEM
 
 | | 6 ALPHA HYDROXYMETHYL PENICILLOIC ACID ACYLATED ON THE TEM-1 BETA-LACTAMASE FROM ESCHERICHIA COLI | | Descriptor: | 2-(1-CARBOXY-2-HYDROXY-ETHYL)-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, TEM-1 BETA LACTAMASE | | Authors: | Maveyraud, L, Massova, I, Samama, J.P, Mobashery, S. | | Deposit date: | 1996-05-28 | | Release date: | 1997-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of 6Alpha-Hydroxymethylpenicillanate Complexed to the Tem-1 Beta-Lactamase from Escherichia Coli: Evidence on the Mechanism of Action of a Novel Inhibitor Designed by a Computer-Aided Process
J.Am.Chem.Soc., 118, 1996
|
|
4K2A
 
 | | Crystal structure of haloalkane dehalogenase DbeA from Bradyrhizobium elkani USDA94 | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Prudnikova, T, Chaloupkova, R, Rezacova, P, Mozga, T, Koudelakova, T, Sato, Y, Kuty, M, Nagata, Y, Damborsky, J, Kuta Smatanova, I, Structure 2 Function Project (S2F) | | Deposit date: | 2013-04-08 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of a novel haloalkane dehalogenase with two halide-binding sites.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1XSM
 
 | | PROTEIN R2 OF RIBONUCLEOTIDE REDUCTASE FROM MOUSE | | Descriptor: | FE (III) ION, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Kauppi, B, Nielsen, B.N, Ramaswamy, S, Kjoller-Larsen, I, Thelander, M, Thelander, L, Eklund, H. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of mammalian ribonucleotide reductase protein R2 reveals a more-accessible iron-radical site than Escherichia coli R2.
J.Mol.Biol., 262, 1996
|
|
4MDC
 
 | | Crystal structure of glutathione S-transferase from Sinorhizobium meliloti 1021, NYSGRC target 021389 | | Descriptor: | GLYCEROL, Putative glutathione S-transferase | | Authors: | Shabalin, I.G, Bacal, P, Cooper, D.R, Stead, M, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-22 | | Release date: | 2013-09-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of glutathione S-transferase from Sinorhizobium meliloti 1021
To be Published
|
|
3IQW
 
 | | AMPPNP complex of C. therm. Get3 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tail-anchored protein targeting factor Get3, ... | | Authors: | Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2009-08-21 | | Release date: | 2009-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into tail-anchored protein binding and membrane insertion by Get3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1SSL
 
 | | Solution structure of the PSI domain from the Met receptor | | Descriptor: | Hepatocyte growth factor receptor | | Authors: | Kozlov, G, Perreault, A, Schrag, J.D, Cygler, M, Gehring, K, Ekiel, I. | | Deposit date: | 2004-03-24 | | Release date: | 2004-10-12 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Insights into function of PSI domains from structure of the Met receptor PSI domain.
Biochem.Biophys.Res.Commun., 321, 2004
|
|
3IRP
 
 | | Crystal structure of functional region of UafA from Staphylococcus saprophyticus at 1.50 angstrom resolution | | Descriptor: | GLYCEROL, POTASSIUM ION, Uro-adherence factor A | | Authors: | Tanaka, Y, Shouji, Y, Matsuoka, E, Kuroda, M, Tanaka, I, Yao, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the functional region of Uro-adherence factor A from Staphylococcus saprophyticus reveals participation of the B domain in ligand binding
Protein Sci., 20, 2011
|
|
4LQK
 
 | | Structure of the vaccinia virus NF- B antagonist A46 | | Descriptor: | BROMIDE ION, Protein A46, SODIUM ION | | Authors: | Grishkovskaya, I, Fedosyuk, S, Skern, T, Djinovic-Carugo, K. | | Deposit date: | 2013-07-18 | | Release date: | 2013-12-25 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Characterization and Structure of the Vaccinia Virus NF-kappa B Antagonist A46.
J.Biol.Chem., 289, 2014
|
|
1SVD
 
 | | The structure of Halothiobacillus neapolitanus RuBisCo | | Descriptor: | GLYCEROL, Ribulose bisphosphate carboxylase small chain, SULFATE ION, ... | | Authors: | Kerfeld, C.A, Sawaya, M.R, Pashkov, I, Cannon, G, Williams, E, Tran, K, Yeates, T.O. | | Deposit date: | 2004-03-29 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Halothiobacillus neapolitanus RuBisCo
To be Published
|
|
3IS5
 
 | | Crystal structure of CDPK kinase domain from toxoplasma Gondii, TGME49_018720 | | Descriptor: | CALCIUM ION, Calcium-dependent protein kinase, GLYCEROL, ... | | Authors: | Wernimont, A.K, Artz, J.D, Senisterra, G, MacKenzie, F, Hutchinson, A, Kozieradzki, I, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-25 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of CDPK kinase domain from toxoplasma Gondii, TGME49_018720
To be Published
|
|
1Z24
 
 | | The molecular structure of insecticyanin from the tobacco hornworm Manduca sexta L. at 2.6 A resolution. | | Descriptor: | BILIVERDIN IX GAMMA CHROMOPHORE, Insecticyanin A form | | Authors: | Holden, H.M, Rypniewski, W.R, Law, J.H, Rayment, I. | | Deposit date: | 2005-03-07 | | Release date: | 2005-04-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The molecular structure of insecticyanin from the tobacco hornworm Manduca sexta L. at 2.6 A resolution.
Embo J., 6, 1987
|
|
3PZC
 
 | | Crystal structure of class II aaRS homologue (Bll0957) complexed with Coenzyme A | | Descriptor: | ACETATE ION, Amino acid--[acyl-carrier-protein] ligase 1, COENZYME A, ... | | Authors: | Weygand-Durasevic, I, Luic, M, Mocibob, M, Ivic, N, Subasic, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate Recognition by Novel Family of Amino Acid:[Carrier Protein] Ligases
Croatica Chemica Acta, 84, 2011
|
|
3B07
 
 | | Crystal structure of octameric pore form of gamma-hemolysin from Staphylococcus aureus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Gamma-hemolysin component A, Gamma-hemolysin component B | | Authors: | Yamashita, K, Kawai, Y, Tanaka, Y, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-06 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Crystal structure of the octameric pore of
staphylococcal gamma-hemolysin reveals the beta-barrel
pore formation mechanism by two components
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3IYA
 
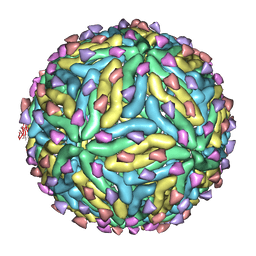 | | Association of the pr peptides with dengue virus blocks membrane fusion at acidic pH | | Descriptor: | Envelope protein, prM protein | | Authors: | Yu, I, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Rossmann, M.G, Chen, J. | | Deposit date: | 2009-06-01 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (22 Å) | | Cite: | Association of the pr peptides with dengue virus at acidic pH blocks membrane fusion.
J.Virol., 83, 2009
|
|
3RRO
 
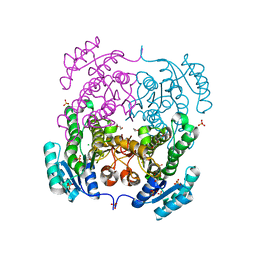 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG) from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, 3-ketoacyl-(acyl-carrier-protein) reductase, CHLORIDE ION, ... | | Authors: | Hou, J, Chruszcz, M, Cooper, D.R, Grabowski, M, Zheng, H, Osinski, T, Shumilin, I, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
4EM9
 
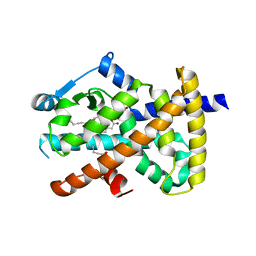 | | Human PPAR gamma in complex with nonanoic acids | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor gamma, ... | | Authors: | Liberato, M.V, Nascimento, A.S, Polikarpov, I. | | Deposit date: | 2012-04-11 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Medium chain fatty acids are selective peroxisome proliferator activated receptor (PPAR) Gamma activators and pan-PPAR partial agonists
Plos One, 7, 2012
|
|
4KE6
 
 | | Crystal structure D196N mutant of Monoglyceride lipase from Bacillus sp. H257 in complex with 1-rac-lauroyl glycerol | | Descriptor: | (2R)-2,3-dihydroxypropyl dodecanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, Thermostable monoacylglycerol lipase | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
1XWJ
 
 | | Vinculin head (1-258) in complex with the talin vinculin binding site 3 (1945-1969) | | Descriptor: | Talin, Vinculin | | Authors: | Gingras, A.R, Papagrigoriou, E, Barsukov, I.L, Critchley, D.R, Emsley, J. | | Deposit date: | 2004-11-01 | | Release date: | 2005-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Dynamic Characterization of a Vinculin Binding Site in the Talin Rod
Biochemistry, 45, 2006
|
|
3EW5
 
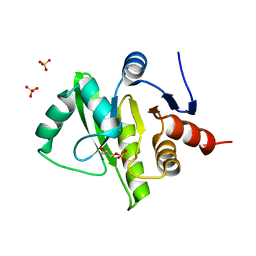 | | Structure of the tetragonal crystal form of X (ADRP) domain from FCoV | | Descriptor: | CHLORIDE ION, SN-GLYCEROL-1-PHOSPHATE, SULFATE ION, ... | | Authors: | Wojdyla, J.A, Manolaridis, I, Tucker, P.A. | | Deposit date: | 2008-10-14 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the X (ADRP) domain of nsp3 from feline coronavirus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1VAG
 
 | | Neuronal nitric oxide synthase oxygenase domain complexed with the inhibitor AR-R17477 | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-(4-{2-[(3-CHLOROBENZYL)AMINO]ETHYL}PHENYL)THIOPHENE-2-CARBOXIMIDAMIDE, Nitric-oxide synthase, ... | | Authors: | Fedorov, R, Vasan, R, Ghosh, D.K, Schlichting, I. | | Deposit date: | 2004-02-16 | | Release date: | 2004-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of nitric oxide synthase isoforms complexed with the inhibitor AR-R17477 suggest a rational basis for specificity and inhibitor design
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
