4E2S
 
 | |
2PS9
 
 | | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli | | Descriptor: | COBALT (II) ION, High-affinity zinc uptake system protein znuA | | Authors: | Yatsunyk, L.A, Kim, L.R, Vorontsov, I.I, Rosenzweig, A.C. | | Deposit date: | 2007-05-04 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli.
J.Biol.Inorg.Chem., 13, 2008
|
|
2PRS
 
 | | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli | | Descriptor: | High-affinity zinc uptake system protein znuA, ISOPROPYL ALCOHOL, ZINC ION | | Authors: | Yatsunyk, L.A, Kim, L.R, Vorontsov, I.I, Rosenzweig, A.C. | | Deposit date: | 2007-05-04 | | Release date: | 2007-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli.
J.Biol.Inorg.Chem., 13, 2008
|
|
2V4J
 
 | | THE CRYSTAL STRUCTURE OF Desulfovibrio vulgaris DISSIMILATORY SULFITE REDUCTASE BOUND TO DsrC PROVIDES NOVEL INSIGHTS INTO THE MECHANISM OF SULFATE RESPIRATION | | Descriptor: | 3,3',3'',3'''-[(1R,2S,3S,4S,7S,8S,11S,12S,13S,16S,19S)-3,8,13,17-tetrakis(carboxylatomethyl)-8,13-dimethyl-1,2,3,4,7,8,11,12,13,16,19,20,22,24-tetradecahydroporphyrin-2,7,12,18-tetrayl]tetrapropanoate, IRON/SULFUR CLUSTER, SIROHEME, ... | | Authors: | Oliveira, T.F, Vonrhein, C, Matias, P.M, Venceslau, S.S, Pereira, I.A.C, Archer, M. | | Deposit date: | 2008-09-22 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Desulfovibrio Vulgaris Dissimilatory Sulfite Reductase Bound to Dsrc Provides Novel Insights Into the Mechanism of Sulfate Respiration.
J.Biol.Chem., 283, 2008
|
|
1XTM
 
 | | Crystal structure of the double mutant Y88H-P104H of a SOD-like protein from Bacillus subtilis. | | Descriptor: | COPPER (II) ION, Hypothetical superoxide dismutase-like protein yojM, ZINC ION | | Authors: | Calderone, V, Mangani, S, Banci, L, Benvenuti, M, Bertini, I, Fantoni, A, Viezzoli, M.S. | | Deposit date: | 2004-10-22 | | Release date: | 2005-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | From an Inactive Prokaryotic SOD Homologue to an Active Protein through Site-Directed Mutagenesis.
J.Am.Chem.Soc., 127, 2005
|
|
4GTB
 
 | | T. Maritima FDTS with FAD, dUMP, and Raltitrexed. | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mathews, I.I, Lesley, S.A, Kohen, A. | | Deposit date: | 2012-08-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Folate binding site of flavin-dependent thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2V63
 
 | | Crystal structure of Rubisco from Chlamydomonas reinhardtii with a large-subunit V331A mutation | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Satagopagan, S, Taylor, T.C, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2007-07-13 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of Altered Large-Subunit Loop-6-Carboxy-Terminus Interactions that Influence Catalytic Efficiency and Co2 O2 Specificity of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase
Biochemistry, 46, 2007
|
|
4C35
 
 | | PKA-S6K1 Chimera with compound 1 (NU1085) bound | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-hydroxyphenyl)-1H-benzimidazole-4-carboxamide, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, ... | | Authors: | Couty, S, Westwood, I.M, Kalusa, A, Cano, C, Travers, J, Boxall, K, Chow, C.L, Burns, S, Schmitt, J, Pickard, L, Barillari, C, McAndrew, P.C, Clarke, P.A, Linardopoulos, S, Griffin, R.J, Aherne, G.W, Raynaud, F.I, Workman, P, Jones, K, van Montfort, R.L.M. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The discovery of potent ribosomal S6 kinase inhibitors by high-throughput screening and structure-guided drug design.
Oncotarget, 4, 2013
|
|
1XEF
 
 | | Crystal structure of the ATP/Mg2+ bound composite dimer of HlyB-NBD | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Alpha-hemolysin translocation ATP-binding protein hlyB, MAGNESIUM ION | | Authors: | Zaitseva, J, Jenewein, S, Holland, I.B, Schmitt, L. | | Deposit date: | 2004-09-10 | | Release date: | 2005-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB
Embo J., 24, 2005
|
|
7EST
 
 | |
2V68
 
 | | Crystal structure of Chlamydomonas reinhardtii Rubisco with large- subunit mutations V331A, T342I | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Satagopan, S, Taylor, T.C, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2007-07-13 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of Altered Large-Subunit Loop-6-Carboxy-Terminus Interactions that Influence Catalytic Efficiency and Co2 O2 Specificity of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase
Biochemistry, 46, 2007
|
|
4C33
 
 | | PKA-S6K1 Chimera Apo | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA | | Authors: | Couty, S, Westwood, I.M, Kalusa, A, Cano, C, Travers, J, Boxall, K, Chow, C.L, Burns, S, Schmitt, J, Pickard, L, Barillari, C, McAndrew, P.C, Clarke, P.A, Linardopoulos, S, Griffin, R.J, Aherne, G.W, Raynaud, F.I, Workman, P, Jones, K, van Montfort, R.L.M. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The discovery of potent ribosomal S6 kinase inhibitors by high-throughput screening and structure-guided drug design.
Oncotarget, 4, 2013
|
|
1XW6
 
 | | 1.9 angstrom resolution structure of human glutathione S-transferase M1A-1A complexed with glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase Mu 1 | | Authors: | Patskovsky, Y, Patskovska, L, Almo, S.C, Listowsky, I. | | Deposit date: | 2004-10-29 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transition state model and mechanism of nucleophilic aromatic substitution reactions catalyzed by human glutathione S-transferase M1a-1a.
Biochemistry, 45, 2006
|
|
1XZO
 
 | | Identification of a disulfide switch in BsSco, a member of the Sco family of cytochrome c oxidase assembly proteins | | Descriptor: | CADMIUM ION, CALCIUM ION, Hypothetical protein ypmQ | | Authors: | Ye, Q, Imriskova-Sosova, I, Hill, B.C, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-11-12 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Identification of a Disulfide Switch in BsSco, a Member of the Sco Family of Cytochrome c Oxidase Assembly Proteins
Biochemistry, 44, 2005
|
|
4NM8
 
 | |
2V67
 
 | | Crystal structure of Chlamydomonas reinhardtii Rubisco with a large- subunit supressor mutation T342I | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Satagopan, S, Taylor, T.C, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2007-07-13 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Altered Large-Subunit Loop-6-Carboxy-Terminus Interactions that Influence Catalytic Efficiency and Co2 O2 Specificity of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase
Biochemistry, 46, 2007
|
|
3ROG
 
 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Thymidine 3'-monophosphate | | Descriptor: | Apolipoprotein A-I-binding protein, SULFATE ION, THYMIDINE-3'-PHOSPHATE | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-25 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
2V27
 
 | | Structure of the cold active phenylalanine hydroxylase from Colwellia psychrerythraea 34H | | Descriptor: | FE (III) ION, PHENYLALANINE HYDROXYLASE, SULFATE ION | | Authors: | Leiros, H.-K.S, Pey, A.L, Innselset, M, Moe, E, Leiros, I, Steen, I.H, Martinez, A. | | Deposit date: | 2007-06-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Phenylalanine Hydroxylase from Colwellia Psychrerythraea 34H, a Monomeric Cold Active Enzyme with Local Flexibility Around the Active Site and High Overall Stability.
J.Biol.Chem., 282, 2007
|
|
2EV4
 
 | | Structure of Rv1264N, the regulatory domain of the mycobacterial adenylyl cylcase Rv1264, with a salt precipitant | | Descriptor: | CHLORIDE ION, Hypothetical protein Rv1264/MT1302, OLEIC ACID | | Authors: | Findeisen, F, Tews, I, Sinning, I. | | Deposit date: | 2005-10-30 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The structure of the regulatory domain of the adenylyl cyclase Rv1264 from Mycobacterium tuberculosis with bound oleic acid
J.Mol.Biol., 369, 2007
|
|
3RNO
 
 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with NADP. | | Descriptor: | Apolipoprotein A-I-binding protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-22 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3ROZ
 
 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Nicotinamide | | Descriptor: | Apolipoprotein A-I-binding protein, NICOTINAMIDE, SULFATE ION | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3AIY
 
 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, REFINED AVERAGE STRUCTURE | | Descriptor: | PHENOL, PROTEIN (INSULIN) | | Authors: | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | Deposit date: | 1998-12-29 | | Release date: | 2000-02-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
4D8P
 
 | | Structural and functional studies of the trans-encoded HLA-DQ2.3 (DQA1*03:01/DQB1*02:01) molecule | | Descriptor: | GLYCEROL, HLA-DQA1 protein, Peptide from Gamma-gliadin,HLA class II histocompatibility antigen, ... | | Authors: | Kim, C.-Y, Hotta, K, Mathews, I.I, Chen, X. | | Deposit date: | 2012-01-11 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural and functional studies of trans-encoded HLA-DQ2.3 (DQA1*03:01/DQB1*02:01) protein molecule
J.Biol.Chem., 287, 2012
|
|
4LLN
 
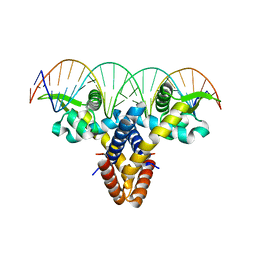 | | Crystal structure of S. aureus MepR-DNA complex | | Descriptor: | MepR, Palindromized mepR operator sequence | | Authors: | Birukou, I, Brennan, R.G. | | Deposit date: | 2013-07-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.842 Å) | | Cite: | Structural mechanism of transcription regulation of the Staphylococcus aureus multidrug efflux operon mepRA by the MarR family repressor MepR.
Nucleic Acids Res., 42, 2014
|
|
2HLC
 
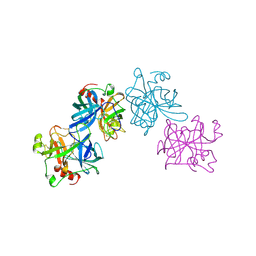 | | HL COLLAGENASE STRUCTURE AT 1.7A RESOLUTION | | Descriptor: | COLLAGENASE | | Authors: | Broutin, I, Merigeau, K, Arnoux, B, Ducruix, A. | | Deposit date: | 1997-01-08 | | Release date: | 1997-09-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.8 A structure of Hypoderma lineatum collagenase: a member of the serine proteinase family.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
