2YGC
 
 | |
4AN9
 
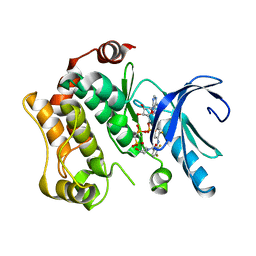 | | Crystal structures of human MEK1 with carboxamide-based allosteric inhibitor XL518 (GDC-0973), or related analogs. | | 分子名称: | DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | 著者 | Rice, K.D, Aay, N, Anand, N.K, Blazey, C.M, Bowles, O.J, Bussenius, J, Costanzo, S, Curtis, J.K, Defina, S.C, Dubenko, L, Engst, S, Joshi, A.A, Kennedy, A.R, Kim, A.I, Koltun, E.S, Lougheed, J.C, Manalo, J.C.L, Martini, J.F, Nuss, J.M, Peto, C.J, Tsang, T.H, Yu, P, Johnston, S. | | 登録日 | 2012-03-16 | | 公開日 | 2012-12-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Novel Carboxamide-Based Allosteric Mek Inhibitors: Discovery and Optimization Efforts Toward Xl518 (Gdc-0973)
Acs Med.Chem.Lett., 3, 2012
|
|
1EKG
 
 | | MATURE HUMAN FRATAXIN | | 分子名称: | FRATAXIN | | 著者 | Dhe-Paganon, S, Shigeta, R, Chi, Y.I, Ristow, M, Shoelson, S.E. | | 登録日 | 2000-03-08 | | 公開日 | 2000-11-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of human frataxin.
J.Biol.Chem., 275, 2000
|
|
1EQT
 
 | | MET-RANTES | | 分子名称: | SULFATE ION, T-CELL SPECIFIC RANTES PROTEIN | | 著者 | Hoover, D.M, Shaw, J, Gryczynski, Z, Proudfoot, A.E.I, Wells, T. | | 登録日 | 2000-04-06 | | 公開日 | 2000-04-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The Crystal Structure of MET-RANTES: Comparison with Native RANTES and AOP-RANTES
PROTEIN PEPT.LETT., 7, 2000
|
|
4RW2
 
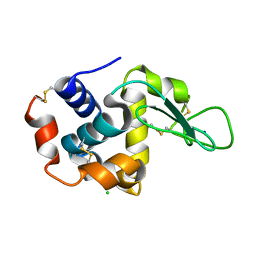 | | Hen egg-white lysozyme structure from a spent-beam experiment at LCLS: refocused beam | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Boutet, S, Foucar, L, Barends, T, Doak, R.B, Koglin, J.E, Messerschmidt, M, Nass, K, Schlichting, I, Shoeman, R, Williams, G.J. | | 登録日 | 2014-12-01 | | 公開日 | 2015-05-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Characterization and use of the spent beam for serial operation of LCLS.
J.SYNCHROTRON RADIAT., 22, 2015
|
|
2B1A
 
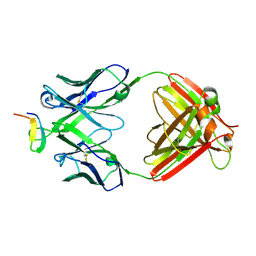 | | Crystal structure analysis of anti-HIV-1 V3 Fab 2219 in complex with UG1033 peptide | | 分子名称: | Fab 2219, heavy chain, light chain, ... | | 著者 | Stanfield, R.L, Gorny, M.K, Zolla-Pazner, S, Wilson, I.A. | | 登録日 | 2005-09-15 | | 公開日 | 2006-07-04 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.348 Å) | | 主引用文献 | Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity.
J.Virol., 80, 2006
|
|
4RS2
 
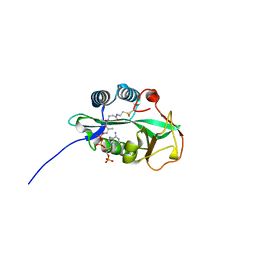 | | 1.55 Angstrom Crystal Structure of GNAT Family N-acetyltransferase (YhbS) from Escherichia coli in Complex with CoA | | 分子名称: | COENZYME A, Predicted acyltransferase with acyl-CoA N-acyltransferase domain | | 著者 | Minasov, G, Wawrzak, Z, Kuhn, M, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-11-06 | | 公開日 | 2014-11-19 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | 1.55 Angstrom Crystal Structure of GNAT Family N-acetyltransferase (YhbS) from Escherichia coli in Complex with CoA.
TO BE PUBLISHED
|
|
4RTD
 
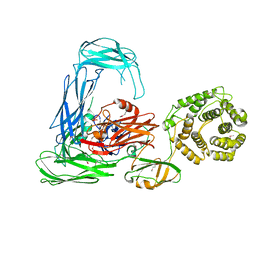 | | Escherichia coli alpha-2-macroglobulin activated by porcine elastase | | 分子名称: | Uncharacterized lipoprotein YfhM | | 著者 | Fyfe, C.D, Grinter, R, Roszak, A.W, Josts, I, Cogdell, R.J, Walker, D. | | 登録日 | 2014-11-14 | | 公開日 | 2015-07-15 | | 最終更新日 | 2015-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.65 Å) | | 主引用文献 | Structure of protease-cleaved Escherichia coli alpha-2-macroglobulin reveals a putative mechanism of conformational activation for protease entrapment.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1E5R
 
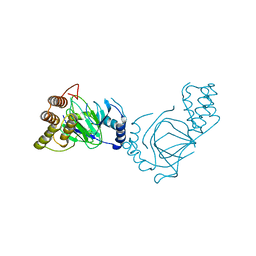 | | Proline 3-hydroxylase (type II) -apo form | | 分子名称: | PROLINE OXIDASE | | 著者 | Clifton, I.J, Hsueh, L.C, Baldwin, J.E, Schofield, C.J, Harlos, K. | | 登録日 | 2000-07-28 | | 公開日 | 2001-07-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of proline 3-hydroxylase. Evolution of the family of 2-oxoglutarate dependent oxygenases.
Eur.J.Biochem., 268, 2001
|
|
1PG6
 
 | | X-Ray Crystal Structure of Protein SPYM3_0169 from Streptococcus pyogenes. Northeast Structural Genomics Consortium Target DR2. | | 分子名称: | CALCIUM ION, Hypothetical protein SpyM3_0169 | | 著者 | Kuzin, A, Lee, I, Edstrom, W, Xiao, R, Acton, T, Rost, B, Montelione, G, Hunt, J, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2003-05-27 | | 公開日 | 2003-12-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | X-ray structure of hypothetical protein SPYM3_0169 from Streptococcus pyogenes
To be Published, 2003
|
|
2B4N
 
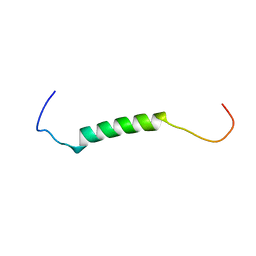 | |
4JMW
 
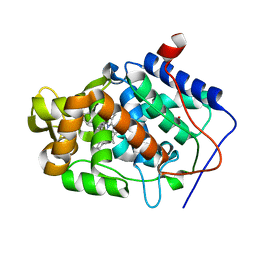 | |
4OJ4
 
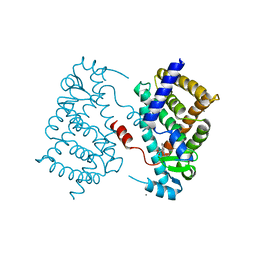 | |
1Q2H
 
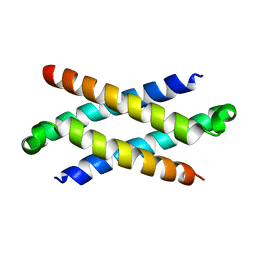 | | Phenylalanine Zipper Mediates APS Dimerization | | 分子名称: | adaptor protein with pleckstrin homology and src homology 2 domains | | 著者 | Dhe-Paganon, S, Werner, E.D, Nishi, M, Chi, Y.-I, Shoelson, S.E. | | 登録日 | 2003-07-24 | | 公開日 | 2004-08-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A phenylalanine zipper mediates APS dimerization.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2BCT
 
 | |
4U14
 
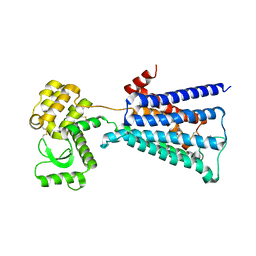 | | Structure of the M3 muscarinic acetylcholine receptor bound to the antagonist tiotropium crystallized with disulfide-stabilized T4 lysozyme (dsT4L) | | 分子名称: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, Muscarinic acetylcholine receptor M3,Endolysin,Muscarinic acetylcholine receptor M3 | | 著者 | Thorsen, T.S, Matt, R.A, Weis, W.I, Kobilka, B.K. | | 登録日 | 2014-07-15 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.57 Å) | | 主引用文献 | Modified T4 Lysozyme Fusion Proteins Facilitate G Protein-Coupled Receptor Crystallogenesis.
Structure, 22, 2014
|
|
1HUU
 
 | |
1PV6
 
 | | Crystal structure of lactose permease | | 分子名称: | Lactose permease | | 著者 | Abramson, J, Smirnova, I, Kasho, V, Verner, G, Kaback, H.R, Iwata, S. | | 登録日 | 2003-06-26 | | 公開日 | 2003-08-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structure and mechanism of the lactose permease of Escherichia coli
SCIENCE, 301, 2003
|
|
2B8I
 
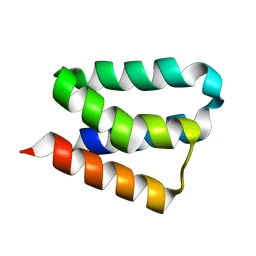 | | Crystal Structure and Functional Studies Reveal that PAS Factor from Vibrio vulnificus is a Novel Member of the Saposin-Fold Family | | 分子名称: | PAS factor | | 著者 | Lee, J.H, Yang, S.T, Rho, S.H, Im, Y.J, Kim, S.Y, Kim, Y.R, Kim, M.K, Kang, G.B, Kim, J.I, Rhee, J.H, Eom, S.H. | | 登録日 | 2005-10-07 | | 公開日 | 2006-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure and functional studies reveal that PAS factor from Vibrio vulnificus is a novel member of the saposin-fold family
J.Mol.Biol., 355, 2006
|
|
3UPD
 
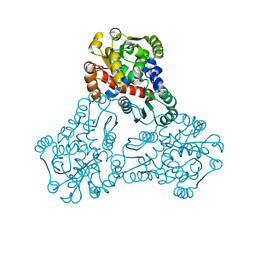 | | 2.9 Angstrom Crystal Structure of Ornithine Carbamoyltransferase (ArgF) from Vibrio vulnificus | | 分子名称: | Ornithine carbamoyltransferase | | 著者 | Minasov, G, Halavaty, A, Shuvalova, L, Winsor, J, Dubrovska, I, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-11-17 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | 2.9 Angstrom Crystal Structure of Ornithine Carbamoyltransferase (ArgF) from Vibrio vulnificus.
TO BE PUBLISHED
|
|
1PWL
 
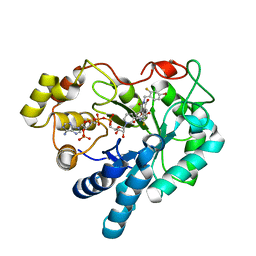 | | Crystal structure of human Aldose Reductase complexed with NADP and Minalrestat | | 分子名称: | 2[4-BROMO-2-FLUOROPHENYL)METHYL]-6-FLUOROSPIRO[ISOQUINOLINE-4-(1H),3'-PYRROLIDINE]-1,2',3,5'(2H)-TETRONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | 著者 | El-Kabbani, O, Darmanin, C, Schneider, T.R, Hazemann, I, Ruiz, F, Oka, M, Joachimiak, A, Schulze-Briese, C, Tomizaki, T, Mitschler, A, Podjarny, A. | | 登録日 | 2003-07-02 | | 公開日 | 2004-02-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Ultrahigh resolution drug design. II. Atomic resolution structures of human aldose reductase holoenzyme complexed with Fidarestat and Minalrestat: implications for the binding of cyclic imide inhibitors
PROTEINS, 55, 2004
|
|
4U12
 
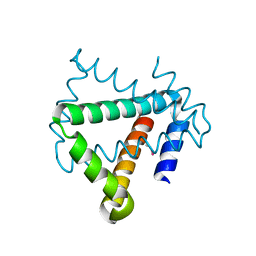 | | Crystal structure of protein HP0242 from Helicobacter pylori at 1.94 A resolution: a knotted homodimer | | 分子名称: | Uncharacterized protein HP0242 | | 著者 | Grabowski, M, Shabalin, I.G, Chruszcz, M, Skarina, T, Onopriyenko, O, Guthrie, J, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-07-14 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Crystal structure of protein HP0242 from Helicobacter pylori at 1.94 A resolution: a knotted homodimer
to be published
|
|
1E9K
 
 | | The structure of the RACK1 interaction sites located within the unique N-terminal region of the cAMP-specific phosphodiesterase, PDE4D5. | | 分子名称: | cAMP-specific 3',5'-cyclic phosphodiesterase 4D | | 著者 | Bolger, G.B, Smith, K.J, McCahill, A, Hyde, E.I, Steele, M.R, Houslay, M.D. | | 登録日 | 2000-10-20 | | 公開日 | 2001-10-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 1H NMR structural and functional characterisation of a cAMP-specific phosphodiesterase-4D5 (PDE4D5) N-terminal region peptide that disrupts PDE4D5 interaction with the signalling scaffold proteins, beta-arrestin and RACK1.
Cell. Signal., 19, 2007
|
|
1PZ4
 
 | | The structural determination of an insect (mosquito) Sterol Carrier Protein-2 with a ligand bound C16 Fatty Acid at 1.35 A resolution | | 分子名称: | PALMITIC ACID, sterol carrier protein 2 | | 著者 | Dyer, D.H, Lovell, S, Thoden, J.B, Holden, H.M, Rayment, I, Lan, Q. | | 登録日 | 2003-07-09 | | 公開日 | 2003-09-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | The Structural Determination of an Insect Sterol Carrier Protein-2 with a Ligand-bound C16 Fatty Acid at 1.35A Resolution
J.Biol.Chem., 278, 2003
|
|
1Z64
 
 | | NMR Solution Structure of Pleurocidin in DPC Micelles | | 分子名称: | Pleruocidin | | 著者 | Syvitski, R.T, Burton, I, Mattatall, N.R, Douglas, S.E, Jakeman, D.L. | | 登録日 | 2005-03-21 | | 公開日 | 2005-04-12 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural characterization of the antimicrobial peptide pleurocidin from winter flounder.
Biochemistry, 44, 2005
|
|
