7OSO
 
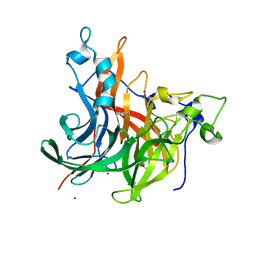 | | The crystal structure of Erwinia tasmaniensis levansucrase in complex with (S)-1,2,4-butanentriol | | 分子名称: | (2~{S})-butane-1,2,4-triol, Levansucrase (Beta-D-fructofuranosyl transferase), ZINC ION | | 著者 | Polsinelli, I, Salomone-Stagni, M, Benini, S. | | 登録日 | 2021-06-09 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Erwinia tasmaniensis levansucrase shows enantiomer selection for (S)-1,2,4-butanetriol.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7OLA
 
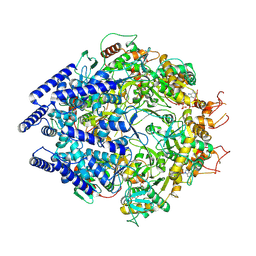 | | Structure of Primase-Helicase in SaPI5 | | 分子名称: | DNA primase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Qiao, C.C, Mir-Sanchis, I. | | 登録日 | 2021-05-19 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Staphylococcal self-loading helicases couple the staircase mechanism with inter domain high flexibility.
Nucleic Acids Res., 50, 2022
|
|
7OM0
 
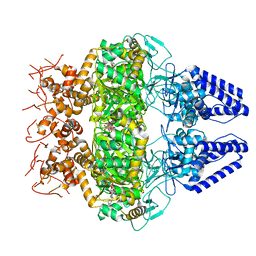 | | Structure of Primase-Helicase in SaPI5 | | 分子名称: | DNA primase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Qiao, C.C, Mir-Sanchis, I. | | 登録日 | 2021-05-21 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Staphylococcal self-loading helicases couple the staircase mechanism with inter domain high flexibility.
Nucleic Acids Res., 50, 2022
|
|
7OJU
 
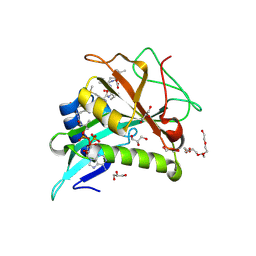 | | Chaetomium thermophilum Naa50 GNAT-domain in complex with bisubstrate analogue CoA-Ac-MVNAL | | 分子名称: | CARBOXYMETHYL COENZYME *A, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | 著者 | Weidenhausen, J, Kopp, J, Sinning, I. | | 登録日 | 2021-05-17 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Extended N-Terminal Acetyltransferase Naa50 in Filamentous Fungi Adds to Naa50 Diversity.
Int J Mol Sci, 23, 2022
|
|
7P3L
 
 | |
8RBX
 
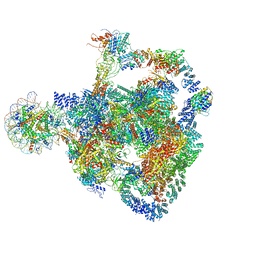 | | Structure of Integrator-PP2A bound to a paused RNA polymerase II-DSIF-NELF-nucleosome complex | | 分子名称: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | 著者 | Fianu, I, Ochmann, M, Walshe, J.L, Cramer, P. | | 登録日 | 2023-12-05 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis of Integrator-dependent RNA polymerase II termination.
Nature, 629, 2024
|
|
2J2M
 
 | | Crystal Structure Analysis of Catalase from Exiguobacterium oxidotolerans | | 分子名称: | CATALASE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Hara, I, Ichise, N, Kojima, K, Kondo, H, Ohgiya, S, Matsuyama, H, Yumoto, I. | | 登録日 | 2006-08-17 | | 公開日 | 2007-01-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Relationship between the Size of the Bottleneck 15 a from Iron in the Main Channel and the Reactivity of Catalase Corresponding to the Molecular Size of Substrates.
Biochemistry, 46, 2007
|
|
1LH1
 
 | | X-RAY STRUCTURAL INVESTIGATION OF LEGHEMOGLOBIN. VI. STRUCTURE OF ACETATE-FERRILEGHEMOGLOBIN AT A RESOLUTION OF 2.0 ANGSTROMS (RUSSIAN) | | 分子名称: | ACETATE ION, LEGHEMOGLOBIN (ACETO MET), PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Vainshtein, B.K, Harutyunyan, E.H, Kuranova, I.P, Borisov, V.V, Sosfenov, N.I, Pavlovsky, A.G, Grebenko, A.I, Konareva, N.V. | | 登録日 | 1982-04-23 | | 公開日 | 1983-01-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-Ray Structural Investigation of Leghemoglobin. Vi. Structure of Acetate-Ferrileghemoglobin at a Resolution of 2.0 Angstroms (Russian)
Kristallografiya, 25, 1980
|
|
7PBW
 
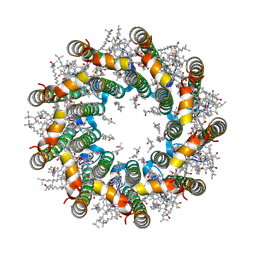 | | Cryo-EM structure of light harvesting complex 2 from Rba. sphaeroides. | | 分子名称: | BACTERIOCHLOROPHYLL A, CALCIUM ION, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | 著者 | Qian, P, Swainsbury, D.J.K, Croll, T.I, Castro-Hartmann, P, Sader, K, Divitini, G, Hunter, C.N. | | 登録日 | 2021-08-02 | | 公開日 | 2021-11-24 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.1 Å) | | 主引用文献 | Cryo-EM Structure of the Rhodobacter sphaeroides Light-Harvesting 2 Complex at 2.1 angstrom.
Biochemistry, 60, 2021
|
|
7PS3
 
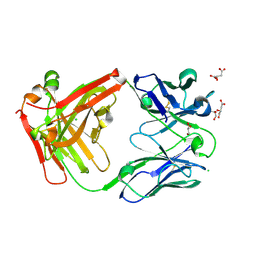 | | Crystal structure of antibody Beta-32 Fab | | 分子名称: | Beta-32 heavy chain, Beta-32 light chain, CHLORIDE ION, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PS0
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with beta-24 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-24 heavy chain, Beta-24 light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PS4
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-38 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-38 Fab heavy chain, Beta-38 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PRZ
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with beta-22 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-22 Fab heavy chain, Beta-22 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PS2
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-29 and Beta-53 Fabs | | 分子名称: | Beta-29 Fab heavy chain, Beta-29 Fab light chain, Beta-53 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PS6
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-44 and Beta-54 Fabs | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, Beta-44 Fab heavy chain, Beta-44 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PIM
 
 | | Partial structure of tyrosine hydroxylase lacking the first 35 residues in complex with dopamine. | | 分子名称: | FE (III) ION, L-DOPAMINE, Regulatory domain alpha-helix, ... | | 著者 | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | 登録日 | 2021-08-20 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
7Q0I
 
 | | Crystal structure of the N-terminal domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-43 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-10-14 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q0G
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-49 and FI-3A Fabs | | 分子名称: | Beta-49 Fab heavy chain, Beta-49 Fab light chain, CHLORIDE ION, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-10-14 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PEN
 
 | |
7PUG
 
 | | GH115 alpha-1,2-glucuronidase in complex with xylopentaose | | 分子名称: | CALCIUM ION, CHLORIDE ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | 著者 | Wilkens, C, Morth, J.P, Polikarpov, I. | | 登録日 | 2021-09-29 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | A GH115 alpha-glucuronidase structure reveals dimerization-mediated substrate binding and a proton wire potentially important for catalysis.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PTM
 
 | | Crystal Structure of Two-Domain Laccase mutant M199G/R240H from Streptomyces griseoflavus | | 分子名称: | COPPER (II) ION, GLYCEROL, OXYGEN MOLECULE, ... | | 著者 | Gabdulkhakov, A, Tishchenko, S, Kolyadenko, I. | | 登録日 | 2021-09-27 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Engineering the Catalytic Properties of Two-Domain Laccase from Streptomyces griseoflavus Ac-993.
Int J Mol Sci, 23, 2021
|
|
7PFR
 
 | |
7PU0
 
 | |
7PES
 
 | | Crystal Structure of Two-Domain Laccase mutant M199G from Streptomyces griseoflavus | | 分子名称: | COPPER (II) ION, OXYGEN MOLECULE, SODIUM ION, ... | | 著者 | Gabdulkhakov, A, Tishchenko, S, Kolyadenko, I. | | 登録日 | 2021-08-11 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Engineering the Catalytic Properties of Two-Domain Laccase from Streptomyces griseoflavus Ac-993.
Int J Mol Sci, 23, 2021
|
|
7PUH
 
 | | Crystal Structure of Two-Domain Laccase mutant H165A/R240H from Streptomyces griseoflavus | | 分子名称: | 1,2-ETHANEDIOL, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kolyadenko, I, Tishchenko, S, Gabdulkhakov, A. | | 登録日 | 2021-09-30 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Engineering the Catalytic Properties of Two-Domain Laccase from Streptomyces griseoflavus Ac-993.
Int J Mol Sci, 23, 2021
|
|
