2WXP
 
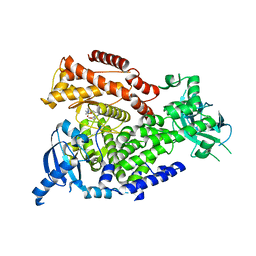 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with GDC-0941. | | 分子名称: | 2-(1H-indazol-4-yl)-6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-yl-thieno[3,2-d]pyrimidine, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | 著者 | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | 登録日 | 2009-11-09 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
1R1K
 
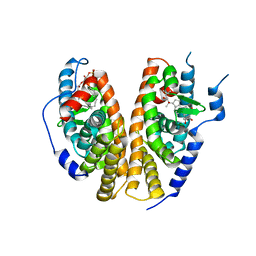 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to ponasterone A | | 分子名称: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone receptor, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, ... | | 著者 | Billas, I.M.L, Iwema, T, Garnier, J.-M, Mitschler, A, Rochel, N, Moras, D, Structural Proteomics in Europe (SPINE) | | 登録日 | 2003-09-24 | | 公開日 | 2003-11-18 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor.
Nature, 426, 2003
|
|
2O0A
 
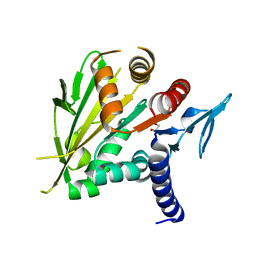 | | The structure of the C-terminal domain of Vik1 has a motor domain fold but lacks a nucleotide-binding site. | | 分子名称: | 1,2-ETHANEDIOL, S.cerevisiae chromosome XVI reading frame ORF YPL253c | | 著者 | Allingham, J.S, Sproul, L.R, Rayment, I, Gilbert, S.P. | | 登録日 | 2006-11-27 | | 公開日 | 2007-03-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Vik1 modulates microtubule-Kar3 interactions through a motor domain that lacks an active site.
Cell(Cambridge,Mass.), 128, 2007
|
|
1R4E
 
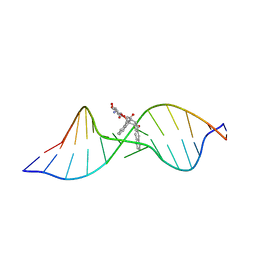 | | Solution structure of the Complex Formed between a Left-Handed Wedge-Shaped Spirocyclic Molecule and Bulged DNA | | 分子名称: | 5'-D(*CP*AP*CP*GP*CP*AP*GP*TP*TP*CP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*GP*AP*TP*GP*CP*GP*TP*G)-3', SPIRO[NAPHTHALENE-2(3H),3'(10'H)-PENTALENO[1,2-B]NAPHTHALENE]-3,10'-DIONE, ... | | 著者 | Hwang, G.S, Jones, G.B, Goldberg, I.H. | | 登録日 | 2003-10-06 | | 公開日 | 2004-04-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Stereochemical control of small molecule binding to bulged DNA: comparison of structures of spirocyclic enantiomer-bulged DNA complexes.
Biochemistry, 43, 2004
|
|
2OBU
 
 | |
6CI4
 
 | | Crystal structure of the formyltransferase PseJ from Anoxybacillus kamchatkensis soaked with UDP-4-amino-4,6-dideoxy-L-AltNAc | | 分子名称: | (2R,3R,4S,5R,6S)-3-(acetylamino)-5-amino-4-hydroxy-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, formyltransferase PseJ | | 著者 | Harb, I, Reimer, J.M, Schmeing, T.M. | | 登録日 | 2018-02-23 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.824068 Å) | | 主引用文献 | Structural Insight into a Novel Formyltransferase and Evolution to a Nonribosomal Peptide Synthetase Tailoring Domain.
ACS Chem. Biol., 13, 2018
|
|
3E8K
 
 | |
1R6H
 
 | | Solution Structure of human PRL-3 | | 分子名称: | protein tyrosine phosphatase type IVA, member 3 isoform 1 | | 著者 | Kozlov, G, Gehring, K, Ekiel, I. | | 登録日 | 2003-10-15 | | 公開日 | 2004-01-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Insights into Molecular Function of the Metastasis-associated Phosphatase PRL-3.
J.Biol.Chem., 279, 2004
|
|
5FDS
 
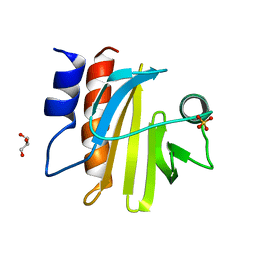 | |
1RH5
 
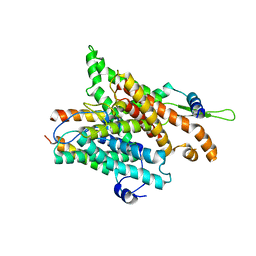 | | The structure of a protein conducting channel | | 分子名称: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, SecBeta | | 著者 | van den Berg, B, Clemons Jr, W.M, Collinson, I, Modis, Y, Hartmann, E, Harrison, S.C, Rapoport, T.A. | | 登録日 | 2003-11-13 | | 公開日 | 2004-01-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | X-ray structure of a protein-conducting channel
Nature, 427, 2004
|
|
5FEF
 
 | |
3ESP
 
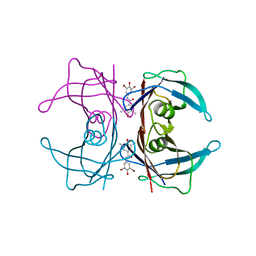 | | Human transthyretin (TTR) complexed with N-(3,5-Dibromo-4-hydroxyphenyl)-3,5-dimethyl-4-hydroxybenzamide | | 分子名称: | N-(3,5-dibromo-4-hydroxyphenyl)-4-hydroxy-3,5-dimethylbenzamide, Transthyretin | | 著者 | Connelly, S, Wilson, I.A. | | 登録日 | 2008-10-06 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Toward optimization of the second aryl substructure common to transthyretin amyloidogenesis inhibitors using biochemical and structural studies.
J.Med.Chem., 52, 2009
|
|
4AHS
 
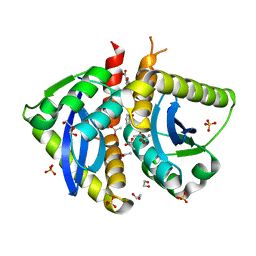 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | 分子名称: | 1,2-ETHANEDIOL, 1-BENZOFURAN-7-CARBOXYLIC ACID, ACETATE ION, ... | | 著者 | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | 登録日 | 2012-02-07 | | 公開日 | 2012-12-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
5O6F
 
 | | NMR structure of cold shock protein A from Corynebacterium pseudotuberculosis | | 分子名称: | Cold-shock protein | | 著者 | Caruso, I.P, Panwalkar, V, Coronado, M.A, Dingley, A.J, Cornelio, M.L, Willbold, D, Arni, R.K, Eberle, R.J. | | 登録日 | 2017-06-06 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and interaction of Corynebacterium pseudotuberculosis cold shock protein A with Y-box single-stranded DNA fragment.
FEBS J., 285, 2018
|
|
3TV3
 
 | |
5FKX
 
 | | Structure of E.coli inducible lysine decarboxylase at active pH | | 分子名称: | LYSINE DECARBOXYLASE, INDUCIBLE | | 著者 | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | 登録日 | 2015-10-20 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.1 Å) | | 主引用文献 | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
5OGU
 
 | | Structure of DNA-binding HU protein from micoplasma Spiroplasma melliferum | | 分子名称: | DNA-binding protein | | 著者 | Altukhov, D.A, Talyzina, A.A, Agapova, Y.K, Vlaskina, A.V, Korzhenevskiy, D.A, Bocharov, E.V, Rakitina, T.V, Timofeev, V.I. | | 登録日 | 2017-07-13 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural plasticity and thermal stability of the histone-like protein from Spiroplasma melliferum are due to phenylalanine insertions into the conservative scaffold.
J.Biomol.Struct.Dyn., 36, 2018
|
|
2O3X
 
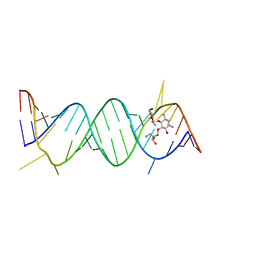 | | Crystal Structure of the Prokaryotic Ribosomal Decoding Site Complexed with Paromamine Derivative NB30 | | 分子名称: | (1R,2R,3S,4R,6S)-4,6-DIAMINO-2-[(5-AMINO-5-DEOXY-BETA-D-RIBOFURANOSYL)OXY]-3-HYDROXYCYCLOHEXYL 2-AMINO-2-DEOXY-ALPHA-D-GLUCOPYRANOSIDE, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | 著者 | Kondo, J, Hainrichson, M, Nudelman, I, Shallom-Shezifi, D, Baasov, T, Westhof, E. | | 登録日 | 2006-12-02 | | 公開日 | 2007-11-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites.
Chembiochem, 8, 2007
|
|
5HQ3
 
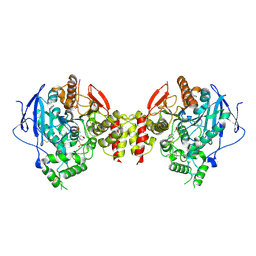 | | Stable, high-expression variant of human acetylcholinesterase | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acetylcholinesterase, O-ETHYLMETHYLPHOSPHONIC ACID ESTER GROUP | | 著者 | Goldenzweig, A, Goldsmith, M, Hill, S.E, Gertman, O, Laurino, P, Ashani, Y, Dym, O, Albeck, S, Unger, T, Prilusky, J, Lieberman, R.L, Aharoni, A, Silman, I, Sussman, J.L, Tawfik, D.S, Fleishman, S.J. | | 登録日 | 2016-01-21 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Automated Structure- and Sequence-Based Design of Proteins for High Bacterial Expression and Stability.
Mol.Cell, 63, 2016
|
|
1V04
 
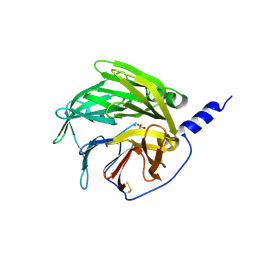 | | serum paraoxonase by directed evolution | | 分子名称: | CALCIUM ION, PHOSPHATE ION, SERUM PARAOXONASE/ARYLESTERASE 1 | | 著者 | Harel, M, Aharoni, A, Gaidukov, L, Brumshtein, B, Khersonsky, O, Yagur, S, Meged, R, Dvir, H, Ravelli, R.B.G, McCarthy, A, Toker, L, Silman, I, Sussman, J.L, Tawfik, D.S. | | 登録日 | 2004-03-22 | | 公開日 | 2004-04-23 | | 最終更新日 | 2017-03-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure and Evolution of the Serum Paraoxonase Family of Detoxifying and Anti-Atherosclerotic Enzymes
Nat.Struct.Mol.Biol., 11, 2004
|
|
5OND
 
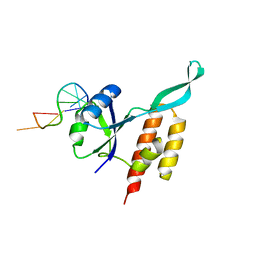 | | RfaH from Escherichia coli in complex with ops DNA | | 分子名称: | DNA (5'-D(*GP*CP*GP*GP*TP*AP*GP*TP*C)-3'), Transcription antitermination protein RfaH | | 著者 | Zuber, P.K, Artsimovitch, I, Roesch, P, Knauer, S.H. | | 登録日 | 2017-08-03 | | 公開日 | 2018-06-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The universally-conserved transcription factor RfaH is recruited to a hairpin structure of the non-template DNA strand.
Elife, 7, 2018
|
|
3U4A
 
 | | From soil to structure: a novel dimeric family 3-beta-glucosidase isolated from compost using metagenomic analysis | | 分子名称: | CALCIUM ION, JMB19063, beta-D-glucopyranose, ... | | 著者 | McAndrew, R.P, Park, J.I, Reindl, W, Friedland, G.D, D'haeseleer, P, Northen, T, Sale, K.L, Simmons, B.A, Adams, P.D. | | 登録日 | 2011-10-07 | | 公開日 | 2013-04-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.196 Å) | | 主引用文献 | From soil to structure: a novel dimeric family 3-beta--glucosidase isolated from compost using metagenomic analysis
To be Published
|
|
6GHA
 
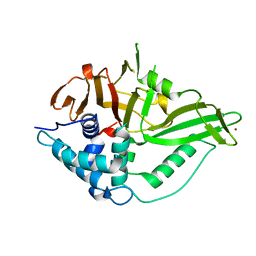 | | USP15 catalytic domain structure | | 分子名称: | Ubiquitin carboxyl-terminal hydrolase 15,Ubiquitin carboxyl-terminal hydrolase 15, ZINC ION | | 著者 | Ward, S.J, Gratton, H.E, Caulton, S.G, Emsley, J, Dreveny, I. | | 登録日 | 2018-05-06 | | 公開日 | 2018-09-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | The structure of the deubiquitinase USP15 reveals a misaligned catalytic triad and an open ubiquitin-binding channel.
J. Biol. Chem., 293, 2018
|
|
3U90
 
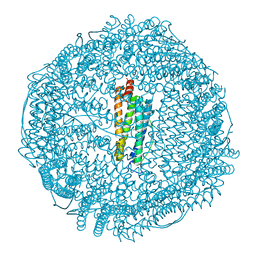 | | apoferritin: complex with SDS | | 分子名称: | CADMIUM ION, DODECYL SULFATE, Ferritin light chain | | 著者 | Liu, R, Bu, W, Xi, J, Mortazavi, S.R, Cheung-Lau, J.C, Dmochowski, I.J, Loll, P.J. | | 登録日 | 2011-10-17 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Beyond the detergent effect: a binding site for sodium dodecyl sulfate (SDS) in mammalian apoferritin.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1UOF
 
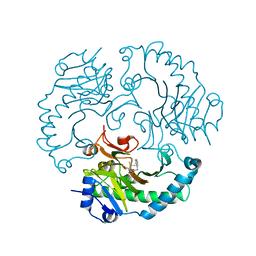 | | Deacetoxycephalosporin C synthase complexed with Penicillin G | | 分子名称: | DEACETOXYCEPHALOSPORIN C SYNTHETASE, FE (II) ION, PENICILLIN G | | 著者 | Valegard, K, Terwisscha Van scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | 登録日 | 2003-09-16 | | 公開日 | 2004-01-09 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
