1GWP
 
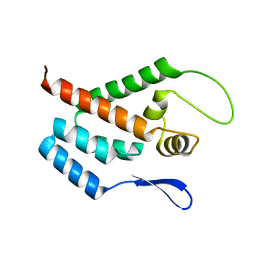 | | STRUCTURE OF THE N-TERMINAL DOMAIN OF THE MATURE HIV-1 CAPSID PROTEIN | | 分子名称: | GAG POLYPROTEIN | | 著者 | Tang, C, Gitti, R.K, Lee, B.M, Walker, J, Summers, M.F, Yoo, S, Sundquist, W.I. | | 登録日 | 2002-03-22 | | 公開日 | 2002-06-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the N-Terminal 283-Residue Fragment of the Immature HIV-1 Gag Polyprotein
Nat.Struct.Biol., 9, 2002
|
|
1AJF
 
 | |
7OO9
 
 | |
1B5P
 
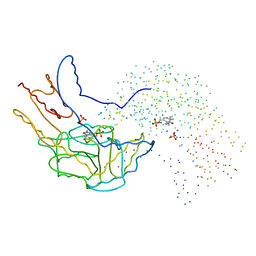 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE DOUBLE MUTANT 1 | | 分子名称: | PHOSPHATE ION, PROTEIN (ASPARTATE AMINOTRANSFERASE), PYRIDOXAL-5'-PHOSPHATE | | 著者 | Ura, H, Nakai, T, Kawaguchi, S.I, Miyahara, I, Hirotsu, K, Kuramitsu, S. | | 登録日 | 1999-01-07 | | 公開日 | 2003-09-02 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Substrate recognition mechanism of thermophilic dual-substrate enzyme
J.BIOCHEM.(TOKYO), 130, 2001
|
|
1BD6
 
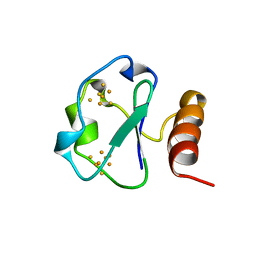 | | 7-FE FERREDOXIN FROM BACILLUS SCHLEGELII, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | 7-FE FERREDOXIN, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER | | 著者 | Aono, S, Bentrop, D, Bertini, I, Donaire, A, Luchinat, C, Niikura, Y, Rosato, A. | | 登録日 | 1998-05-06 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the oxidized Fe7S8 ferredoxin from the thermophilic bacterium Bacillus schlegelii by 1H NMR spectroscopy.
Biochemistry, 37, 1998
|
|
1AQS
 
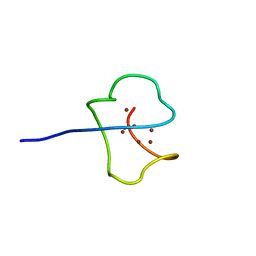 | | CU-METALLOTHIONEIN FROM SACCHAROMYCES CEREVISIAE, NMR, 10 STRUCTURES | | 分子名称: | COPPER (I) ION, CU-METALLOTHIONEIN | | 著者 | Peterson, C.W, Narula, S.S, Armitage, I.M. | | 登録日 | 1997-07-31 | | 公開日 | 1997-12-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 3D solution structure of copper and silver-substituted yeast metallothioneins.
FEBS Lett., 379, 1996
|
|
1B5O
 
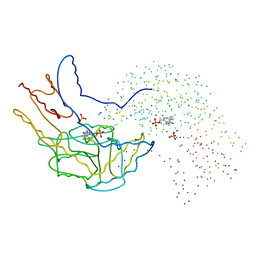 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE SINGLE MUTANT 1 | | 分子名称: | PHOSPHATE ION, PROTEIN (ASPARTATE AMINOTRANSFERASE), PYRIDOXAL-5'-PHOSPHATE | | 著者 | Ura, H, Nakai, T, Kawaguchi, S.I, Miyahara, I, Hirotsu, K, Kuramitsu, S. | | 登録日 | 1999-01-07 | | 公開日 | 2003-09-02 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Substrate recognition mechanism of thermophilic dual-substrate enzyme
J.BIOCHEM.(TOKYO), 130, 2001
|
|
1AX9
 
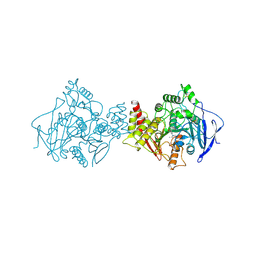 | | ACETYLCHOLINESTERASE COMPLEXED WITH EDROPHONIUM, LAUE DATA | | 分子名称: | ACETYLCHOLINESTERASE, EDROPHONIUM ION | | 著者 | Raves, M.L, Ravelli, R.B.G, Sussman, J.L, Harel, M, Silman, I. | | 登録日 | 1997-11-03 | | 公開日 | 1998-02-11 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Static Laue diffraction studies on acetylcholinesterase.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AQR
 
 | | CU-METALLOTHIONEIN FROM SACCHAROMYCES CEREVISIAE, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | COPPER (I) ION, CU-METALLOTHIONEIN | | 著者 | Peterson, C.W, Narula, S.S, Armitage, I.M. | | 登録日 | 1997-07-31 | | 公開日 | 1997-12-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 3D solution structure of copper and silver-substituted yeast metallothioneins.
FEBS Lett., 379, 1996
|
|
6ZOO
 
 | | Photosystem I reduced Plastocyanin Complex | | 分子名称: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | 著者 | Nelson, N, Caspy, I, Shkolnisky, Y. | | 登録日 | 2020-07-07 | | 公開日 | 2021-06-16 | | 最終更新日 | 2021-07-07 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Structure of plant photosystem I-plastocyanin complex reveals strong hydrophobic interactions.
Biochem.J., 478, 2021
|
|
1BC6
 
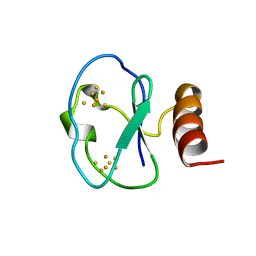 | | 7-FE FERREDOXIN FROM BACILLUS SCHLEGELII, NMR, 20 STRUCTURES | | 分子名称: | 7-FE FERREDOXIN, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER | | 著者 | Aono, S, Bentrop, D, Bertini, I, Donaire, A, Luchinat, C, Niikura, Y, Rosato, A. | | 登録日 | 1998-05-05 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the oxidized Fe7S8 ferredoxin from the thermophilic bacterium Bacillus schlegelii by 1H NMR spectroscopy.
Biochemistry, 37, 1998
|
|
1BDD
 
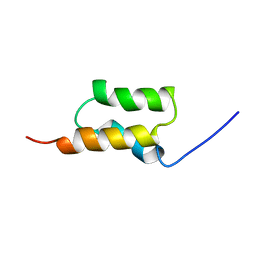 | | STAPHYLOCOCCUS AUREUS PROTEIN A, IMMUNOGLOBULIN-BINDING B DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | STAPHYLOCOCCUS AUREUS PROTEIN A | | 著者 | Gouda, H, Torigoe, H, Saito, A, Sato, M, Arata, Y, Shimada, I. | | 登録日 | 1996-06-28 | | 公開日 | 1997-01-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional solution structure of the B domain of staphylococcal protein A: comparisons of the solution and crystal structures.
Biochemistry, 31, 1992
|
|
7SYX
 
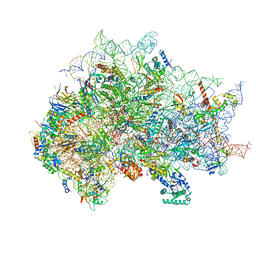 | | Structure of the delta dII IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S24, 40S ribosomal protein S25, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-03-08 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYW
 
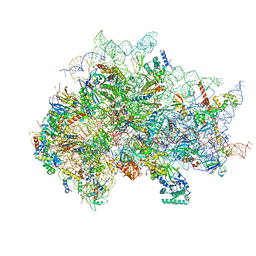 | | Structure of the wt IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(wt) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYU
 
 | | Structure of the delta dII IRES w/o eIF2 48S initiation complex, closed conformation. Structure 13(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
6ZZY
 
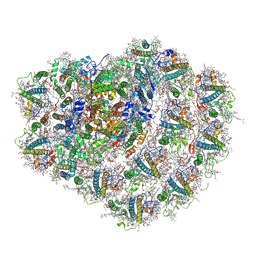 | | Structure of high-light grown Chlorella ohadii photosystem I | | 分子名称: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ... | | 著者 | Caspy, I, Nelson, N, Nechushtai, R, Shkolnisky, Y, Neumann, E. | | 登録日 | 2020-08-05 | | 公開日 | 2021-07-28 | | 最終更新日 | 2021-09-29 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
7A4P
 
 | | Structure of small high-light grown Chlorella ohadii photosystem I | | 分子名称: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (3R)-beta,beta-caroten-3-ol, ... | | 著者 | Caspy, I, Nelson, N, Nechushtai, R, Shkolnisky, Y, Neumann, E. | | 登録日 | 2020-08-20 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
6ZZX
 
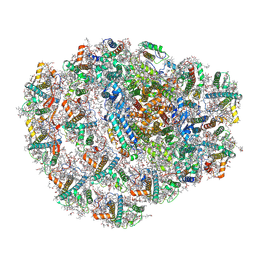 | | Structure of low-light grown Chlorella ohadii Photosystem I | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (3R)-beta,beta-caroten-3-ol, ... | | 著者 | Caspy, I, Nelson, N, Nechushtai, R, Neumann, E, Shkolnisky, Y. | | 登録日 | 2020-08-05 | | 公開日 | 2021-07-28 | | 最終更新日 | 2021-09-29 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
6W1T
 
 | | RT XFEL structure of Photosystem II 250 microseconds after the second illumination at 2.01 Angstrom resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | 登録日 | 2020-03-04 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1BDC
 
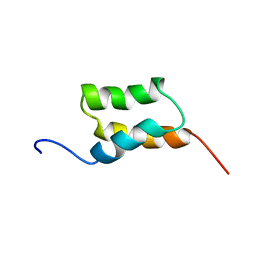 | | STAPHYLOCOCCUS AUREUS PROTEIN A, IMMUNOGLOBULIN-BINDING B DOMAIN, NMR, 10 STRUCTURES | | 分子名称: | STAPHYLOCOCCUS AUREUS PROTEIN A | | 著者 | Gouda, H, Torigoe, H, Saito, A, Sato, M, Arata, Y, Shimada, I. | | 登録日 | 1996-06-28 | | 公開日 | 1997-01-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional solution structure of the B domain of staphylococcal protein A: comparisons of the solution and crystal structures.
Biochemistry, 31, 1992
|
|
1BS8
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | 分子名称: | PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS5
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM | | 分子名称: | PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ZINC ION | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
7SYT
 
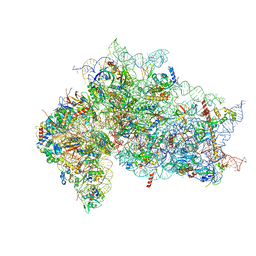 | | Structure of the wt IRES w/o eIF2 48S initiation complex, closed conformation. Structure 13(wt) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYV
 
 | | Structure of the wt IRES eIF5B-containing pre-48S initiation complex, open conformation. Structure 14(wt) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYS
 
 | | Structure of the delta dII IRES eIF2-containing 48S initiation complex, closed conformation. Structure 12(delta dII). | | 分子名称: | 18S rRNA, Eukaryotic translation initiation factor 1A, X-chromosomal, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-27 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
