3EPB
 
 | | Human AdoMetDC E256Q mutant complexed with putrescine | | 分子名称: | 1,4-DIAMINOBUTANE, PYRUVIC ACID, S-adenosylmethionine decarboxylase alpha chain, ... | | 著者 | Bale, S, Lopez, M.M, Makhatadze, G.I, Fang, Q, Pegg, A.E, Ealick, S.E. | | 登録日 | 2008-09-29 | | 公開日 | 2008-12-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Basis for Putrescine Activation of Human S-Adenosylmethionine Decarboxylase.
Biochemistry, 47, 2008
|
|
1Q8O
 
 | | Pterocartpus angolensis lectin PAL in complex with the dimmanoside Man(alpha1-2)Man | | 分子名称: | CALCIUM ION, MANGANESE (II) ION, alpha-D-mannopyranose-(1-2)-methyl alpha-D-mannopyranoside, ... | | 著者 | Loris, R, Van Walle, I, De Greve, H, Beeckmans, S, Deboeck, F, Wyns, L, Bouckaert, J. | | 登録日 | 2003-08-22 | | 公開日 | 2004-02-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Basis of Oligomannose Recognition by the Pterocarpus angolensis Seed Lectin
J.Mol.Biol., 335, 2004
|
|
2YGY
 
 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in space group P21 crystal form II | | 分子名称: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE, PENTAETHYLENE GLYCOL | | 著者 | Campeotto, I, Nelson, A, Berry, A, Phillips, S.E.V, Pearson, A.R. | | 登録日 | 2011-04-23 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Pathological macromolecular crystallographic data affected by twinning, partial-disorder and exhibiting multiple lattices for testing of data processing and refinement tools.
Sci Rep, 8, 2018
|
|
6B9O
 
 | | Structure of GH 38 Jack Bean alpha-mannosidase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-mannosidase from Canavalia ensiformis (jack bean), ZINC ION, ... | | 著者 | Howard, E, Cousido-Siah, A, Lepage, M, Bodlenner, A, Mitschler, A, Meli, A, De Riccardis, F, Izzo, I, Podjarny, A, Compain, P. | | 登録日 | 2017-10-11 | | 公開日 | 2018-09-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.841 Å) | | 主引用文献 | Structural Basis of Outstanding Multivalent Effects in Jack Bean alpha-Mannosidase Inhibition.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6BDH
 
 | | Crystal structure of human CYP3A4 bound to an inhibitor | | 分子名称: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-{[(2S)-3-oxo-2-(phenylamino)-3-{[(pyridin-3-yl)methyl]amino}propyl]sulfanyl}-3-phenylpropan-2-yl]carbamate | | 著者 | Sevrioukova, I. | | 登録日 | 2017-10-23 | | 公開日 | 2017-12-13 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Interaction of the rationally designed ritonavir-like inhibitors with human cytochrome P450 3A4: Impact of the side group interplay
Mol. Pharm., 2017
|
|
2YKS
 
 | |
7NFY
 
 | | P1a-state of wild type human mitochondrial LONP1 protease with bound substrate protein and ATPgS | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial, ... | | 著者 | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | 登録日 | 2021-02-08 | | 公開日 | 2021-02-24 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
3RO7
 
 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Thymine. | | 分子名称: | Apolipoprotein A-I-binding protein, SULFATE ION, THYMINE | | 著者 | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | 登録日 | 2011-04-25 | | 公開日 | 2012-07-18 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
9BLG
 
 | | Crystal structure of non-receptor protein tyrosine phosphatase SHP2 in complex with PF-07284892 | | 分子名称: | (1S)-1'-{6-[(2-amino-3-chloropyridin-4-yl)sulfanyl]-1,2,4-triazin-3-yl}-1,3-dihydrospiro[indene-2,4'-piperidin]-1-amine, Tyrosine-protein phosphatase non-receptor type 11 | | 著者 | Bester, S.M, Wu, W.-I, Mou, T.-C. | | 登録日 | 2024-04-30 | | 公開日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | SHP2 Inhibition Sensitizes Diverse Oncogene-Addicted Solid Tumors to Re-treatment with Targeted Therapy.
Cancer Discov, 13, 2023
|
|
7NG5
 
 | | P1c-state of wild type human mitochondrial LONP1 protease with bound substrate protein in presence of ATP/ADP mix | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | 著者 | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | 登録日 | 2021-02-08 | | 公開日 | 2021-02-24 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
7NG4
 
 | | P1b-state of wild type human mitochondrial LONP1 protease with bound endogenous substrate protein and in presence of ATP/ADP mix | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | 著者 | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | 登録日 | 2021-02-08 | | 公開日 | 2021-02-24 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
3R5L
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Deazaflavin-dependent nitroreductase | | 著者 | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayyar, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | 登録日 | 2011-03-18 | | 公開日 | 2012-01-18 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
6KAY
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW7647 co-crystals obtained by soaking | | 分子名称: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | 登録日 | 2019-06-24 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.735 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB3
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW7647 co-crystals obtained by delipidation and cross-seeding | | 分子名称: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | 登録日 | 2019-06-24 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
3EP4
 
 | | Human AdoMetDC E256Q mutant with no putrescine bound | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYRUVIC ACID, S-adenosylmethionine decarboxylase alpha chain, ... | | 著者 | Bale, S, Lopez, M.M, Makhatadze, G.I, Fang, Q, Pegg, A.E, Ealick, S.E. | | 登録日 | 2008-09-29 | | 公開日 | 2008-12-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structural Basis for Putrescine Activation of Human S-Adenosylmethionine Decarboxylase.
Biochemistry, 47, 2008
|
|
2GJZ
 
 | |
7JU7
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with masitinib | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-08-19 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
2GSY
 
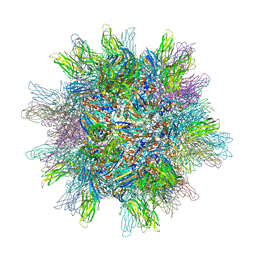 | | The 2.6A structure of Infectious Bursal Virus Derived T=1 Particles | | 分子名称: | CALCIUM ION, polyprotein | | 著者 | Garriga, D, Querol-Audi, J, Abaitua, F, Saugar, I, Pous, J, Verdaguer, N, Caston, J.R, Rodriguez, J.F. | | 登録日 | 2006-04-27 | | 公開日 | 2006-07-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The 2.6-angstrom structure of infectious bursal disease virus-derived t=1 particles reveals new stabilizing elements of the virus capsid.
J.Virol., 80, 2006
|
|
6AOR
 
 | |
7NQZ
 
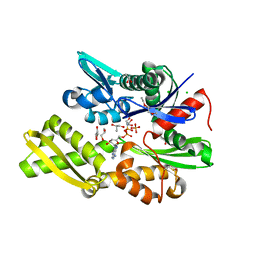 | | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with Z1827898537 | | 分子名称: | (3S)-N-benzylpyrrolidin-3-amine, AMP PHOSPHORAMIDATE, CHLORIDE ION, ... | | 著者 | Mohamad, N, O'Donoghue, A, Kantsadi, A.L, Vakonakis, I. | | 登録日 | 2021-03-02 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.319 Å) | | 主引用文献 | Structures of P. falciparum Hsp70-x nucleotide binding domain with small molecule ligands
To Be Published
|
|
7NQR
 
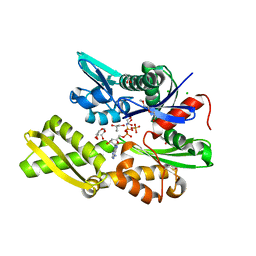 | | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with Z287256168 | | 分子名称: | (S)-N-(1-cyclopropylethyl)-6-methylpicolinamide, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, AMP PHOSPHORAMIDATE, ... | | 著者 | Mohamad, N, O'Donoghue, A, Kantsadi, A.L, Vakonakis, I. | | 登録日 | 2021-03-02 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Structures of P. falciparum Hsp70-x nucleotide binding domain with small molecule ligands
To Be Published
|
|
2IID
 
 | | Structure of L-amino acid oxidase from Calloselasma rhodostoma in complex with L-phenylalanine | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, L-amino-acid oxidase, ... | | 著者 | Moustafa, I.M, Foster, S, Lyubimov, A.Y, Vrielink, A. | | 登録日 | 2006-09-27 | | 公開日 | 2006-10-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of LAAO from Calloselasma rhodostoma with an L-Phenylalanine Substrate: Insights into Structure and Mechanism
J.Mol.Biol., 364, 2006
|
|
7NQU
 
 | | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with Z396380540 | | 分子名称: | AMP PHOSPHORAMIDATE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Mohamad, N, O'Donoghue, A, Kantsadi, A.L, Vakonakis, I. | | 登録日 | 2021-03-02 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Structures of P. falciparum Hsp70-x nucleotide binding domain with small molecule ligands
To Be Published
|
|
7NQS
 
 | | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with Z1203107138 | | 分子名称: | 1-ethyl-N-[(4-fluorophenyl)methyl]-1H-pyrazole-4-carboxamide, AMP PHOSPHORAMIDATE, GLYCEROL, ... | | 著者 | Mohamad, N, O'Donoghue, A, Kantsadi, A.L, Vakonakis, I. | | 登録日 | 2021-03-02 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Structures of P. falciparum Hsp70-x nucleotide binding domain with small molecule ligands
To Be Published
|
|
6AW9
 
 | | 2.55A resolution structure of SAH bound catechol O-methyltransferase (COMT) L136M from Nannospalax galili | | 分子名称: | CALCIUM ION, Catechol O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | 登録日 | 2017-09-05 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
