3TNJ
 
 | | Crystal structure of universal stress protein from Nitrosomonas europaea with AMP bound | | Descriptor: | ADENOSINE MONOPHOSPHATE, Universal stress protein (Usp) | | Authors: | Tkaczuk, K.L, Chruszcz, M, Shumilin, I.A, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-01 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
4AEK
 
 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens | | Descriptor: | ENDOGLUCANASE CEL5A | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-11 | | Release date: | 2013-01-16 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan.
J.Biol.Chem., 288, 2013
|
|
3QK6
 
 | | Crystal structure of Escherichia coli PhnD | | Descriptor: | PhnD, subunit of alkylphosphonate ABC transporter, UNKNOWN LIGAND | | Authors: | Alicea, I, Schreiter, E.R. | | Deposit date: | 2011-01-31 | | Release date: | 2011-10-12 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Escherichia coli Phosphonate Binding Protein PhnD and Rationally Optimized Phosphonate Biosensors.
J.Mol.Biol., 414, 2011
|
|
3V2B
 
 | | Human poly(adp-ribose) polymerase 15 (ARTD7, BAL3), macro domain 2 in complex with adenosine-5-diphosphoribose | | Descriptor: | Poly [ADP-ribose] polymerase 15, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karlberg, T, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Kotzcsh, A, Kraulis, P, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van den berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-12 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains.
Structure, 21, 2013
|
|
3V4G
 
 | | 1.60 Angstrom resolution crystal structure of an arginine repressor from Vibrio vulnificus CMCP6 | | Descriptor: | Arginine repressor | | Authors: | Halavaty, A.S, Minasov, G, Filippova, E, Shuvalova, L, Winsor, J, Dubrovska, I, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.60 Angstrom resolution crystal structure of an arginine repressor from Vibrio vulnificus CMCP6
To be Published
|
|
3QKY
 
 | | Crystal structure of Rhodothermus marinus BamD | | Descriptor: | Outer membrane assembly lipoprotein YfiO | | Authors: | Sandoval, C.M, Baker, S.L, Jansen, K, Metzner, S.I, Sousa, M.C. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of BamD: an essential component of the beta-Barrel assembly machinery of gram-negative bacteria.
J.Mol.Biol., 409, 2011
|
|
3QL3
 
 | | Re-refined coordinates for PDB entry 1RX2 | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
3QLB
 
 | | Enantiopyochelin outer membrane TonB-dependent transporter from Pseudomonas fluorescens bound to the ferri-enantiopyochelin | | Descriptor: | CITRIC ACID, ENANTIO-PYOCHELIN FE(III), Enantio-pyochelin receptor, ... | | Authors: | Brillet, K, Noel, S, Mislin, G.L.A, Reimmann, C, Schalk, I.J, Cobessi, D. | | Deposit date: | 2011-02-02 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Pyochelin enantiomers and their outer-membrane siderophore transporters in fluorescent pseudomonads: structural bases for unique enantiospecific recognition
J.Am.Chem.Soc., 133, 2011
|
|
3V4H
 
 | | Crystal structure of a type VI secretion system effector from Yersinia pestis | | Descriptor: | hypothetical protein | | Authors: | Filippova, E.V, Halavaty, A, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a type VI secretion system effector from Yersinia pestis
To be Published
|
|
3V68
 
 | |
3UKD
 
 | | UMP/CMP KINASE FROM SLIME MOLD COMPLEXED WITH ADP, CMP, AND ALF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Schlichting, I, Reinstein, J. | | Deposit date: | 1997-05-20 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of active conformations of UMP kinase from Dictyostelium discoideum suggest phosphoryl transfer is associative.
Biochemistry, 36, 1997
|
|
4AJW
 
 | | Discovery and Optimization of New Benzimidazole- and Benzoxazole-Pyrimidone Selective PI3KBeta Inhibitors for the Treatment of Phosphatase and TENsin homologue (PTEN)-Deficient Cancers | | Descriptor: | 2-[(1-methyl-1H-benzimidazol-2-yl)methyl]-6-morpholin-4-ylpyrimidin-4(3H)-one, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Certal, V, Halley, F, Virone-Oddos, A, Delorme, C, Karlsson, A, Rak, A, Thompson, F, Filoche-Romme, B, El-Ahmad, Y, Carry, J.C, Abecassis, P.Y, Lejeune, P, Bonnevaux, H, Nicolas, J.P, Bertrand, T, Marquette, J.P, Michot, N, Benard, T, Below, P, Vade, I, Chatreaux, F, Lebourg, G, Pilorge, F, Angouillant-Boniface, O, Louboutin, A, Lengauer, C, Schio, L. | | Deposit date: | 2012-02-20 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Optimization of New Benzimidazole- and Benzoxazole-Pyrimidone Selective Pi3Kbeta Inhibitors for the Treatment of Phosphatase and Tensin Homologue (Pten)-Deficient Cancers.
J.Med.Chem., 55, 2012
|
|
3RE4
 
 | |
4AIX
 
 | | Crystallographic structure of an amyloidogenic variant, 3rC34Y, of the germinal line lambda 3 | | Descriptor: | IG LAMBDA CHAIN V-IV REGION BAU | | Authors: | Villalba, M.I, Luna, O.D, Rudino-Pinera, E, Sanchez, R, Sanchez-Lopez, R, Rojas-Trejo, S, Olamendi-Portugal, T, Fernandez-Velasco, D.A, Becerril, B. | | Deposit date: | 2012-02-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Site-Directed Mutagenesis Reveals Regions Implicated in the Stability and Fiber Formation of Human Lambda3R Light Chains.
J.Biol.Chem., 290, 2015
|
|
4AOF
 
 | | Selective small molecule inhibitor discovered by chemoproteomic assay platform reveals regulation of Th17 cell differentiation by PI3Kgamma | | Descriptor: | N-[6-(5-methylsulfonylpyridin-3-yl)-[1,2,4]triazolo[1,5-a]pyridin-2-yl]ethanamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Bergamini, G, Bell, K, Shimamura, S, Werner, T, Cansfield, A, Muller, K, Perrin, J, Rau, C, Ellard, K, Hopf, C, Doce, C, Leggate, D, Mangano, R, Mathieson, T, OMahony, A, Plavec, I, Rharbaoui, F, Reinhard, F, Savitski, M.M, Ramsden, N, Hirsch, E, Drewes, G, Rausch, O, Bantscheff, M, Neubauer, G. | | Deposit date: | 2012-03-26 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A Selective Inhibitor Reveals Pi3Kgamma Dependence of T(H)17 Cell Differentiation.
Nat.Chem.Biol., 8, 2012
|
|
4AR2
 
 | | Dodecahedron formed of penton base protein from adenovirus Ad3 | | Descriptor: | CALCIUM ION, FIBER PROTEIN, L2 PROTEIN III (PENTON BASE) | | Authors: | Burmeister, W.P, Szolajska, E, Zochowska, M, Nerlo, B, Andreev, I, Schoehn, G, Andrieu, J.-P, Fender, P, Naskalska, A, Zubieta, C, Cusack, S, Chroboczek, J. | | Deposit date: | 2012-04-20 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Structural Basis for the Integrity of Adenovirus Ad3 Dodecahedron.
Plos One, 7, 2012
|
|
3VIM
 
 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with a new glucopyranosidic product | | Descriptor: | 2-{4-[2-(beta-D-glucopyranosyloxy)ethyl]piperazin-1-yl}ethanesulfonic acid, Beta-glucosidase, CHLORIDE ION, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TPK
 
 | | Crystal structure of the oligomer-specific KW1 antibody fragment | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, Immunoglobulin heavy chain antibody variable domain KW1 | | Authors: | Parthier, C, Morgado, I, Stubbs, M.T, Fandrich, M. | | Deposit date: | 2011-09-08 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis of beta-amyloid oligomer recognition with a conformational antibody fragment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VO9
 
 | | Staphylococcus aureus FtsZ apo-form (SeMet) | | Descriptor: | Cell division protein FtsZ | | Authors: | Matsui, T, Yamane, J, Mogi, N, Yao, M, Tanaka, I. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-29 | | Last modified: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VBZ
 
 | | Crystal structure of Taipoxin beta subunit isoform 2 | | Descriptor: | Phospholipase A2 homolog, taipoxin beta chain | | Authors: | Cendron, L, Micetic, I, Polverino, P, Beltramini, M, Paoli, M. | | Deposit date: | 2012-01-03 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural analysis of trimeric phospholipase A(2) neurotoxin from the Australian taipan snake venom.
Febs J., 279, 2012
|
|
3U2I
 
 | | X-ray crystal structure of human Transthyretin at room temperature | | Descriptor: | Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M, Nabeshima, Y, Kusaka, K, Yamada, T, Hosoya, T, Ohhara, T, Kurihara, K, Tomoyori, K, Tanaka, I, Niimura, N. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin
J.Struct.Biol., 177, 2012
|
|
3VIG
 
 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with 1-deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-glucosidase, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U3R
 
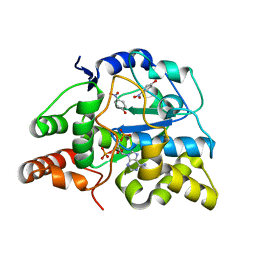 | | Crystal structure of D249G mutated Human SULT1A1 bound to PAP and P-NITROPHENOL | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, P-NITROPHENOL, Sulfotransferase 1A1 | | Authors: | Guttman, C, Berger, I, Aharoni, A, Zarivach, R. | | Deposit date: | 2011-10-06 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The molecular basis for the broad substrate specificity of human sulfotransferase 1A1.
Plos One, 6, 2011
|
|
3VZR
 
 | |
3TM4
 
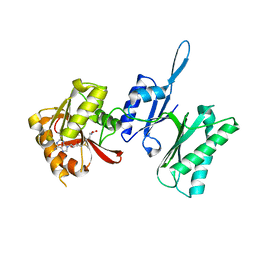 | | Crystal structure of Trm14 from Pyrococcus furiosus in complex with S-adenosylmethionine | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (guanine N2-)-methyltransferase Trm14 | | Authors: | Fislage, M, Roovers, M, Tuszynska, I, Bujnicki, J.M, Droogmans, L, Versees, W. | | Deposit date: | 2011-08-31 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of the tRNA:m2G6 methyltransferase Trm14/TrmN from two domains of life.
Nucleic Acids Res., 40, 2012
|
|
