1N7K
 
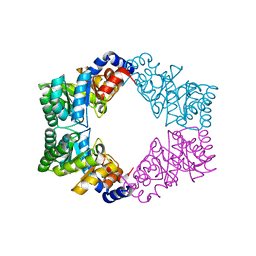 | | Unique tetrameric structure of deoxyribose phosphate aldolase from Aeropyrum pernix | | Descriptor: | deoxyribose-phosphate aldolase | | Authors: | Tsuge, H, Sakuraba, H, Shimoya, I, Katunuma, N, Ago, H, Miyano, M, Ohshima, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-15 | | Release date: | 2003-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The First Crystal Structure of Archaeal Aldolase. UNIQUE TETRAMERIC STRUCTURE of 2-DEOXY-D-RIBOSE-5-PHOSPHATE ALDOLASE FROM THE HYPERTHERMOPHILIC ARCHAEA Aeropyrum pernix.
J.Biol.Chem., 278, 2003
|
|
4GZX
 
 | |
3JV9
 
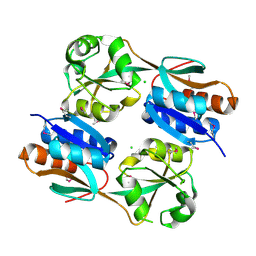 | | The structure of a reduced form of OxyR from N. meningitidis | | Descriptor: | CHLORIDE ION, Transcriptional regulator, LysR family | | Authors: | Sainsbury, S, Ren, J, Stuart, D.I, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-09-16 | | Release date: | 2010-06-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The structure of a reduced form of OxyR from Neisseria meningitidis
Bmc Struct.Biol., 10, 2010
|
|
4GZS
 
 | | N2 neuraminidase D151G mutant of a/Tanzania/205/2010 H3N2 in complex with hepes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Influenza virus neuraminidases with reduced enzymatic activity that avidly bind sialic Acid receptors.
J.Virol., 86, 2012
|
|
7KEG
 
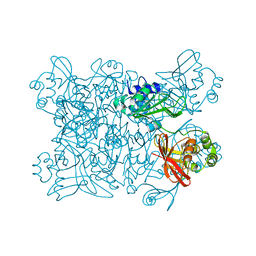 | | Crystal structure from SARS-COV2 NendoU NSP15 | | Descriptor: | PHOSPHATE ION, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | Deposit date: | 2020-10-10 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
1N9N
 
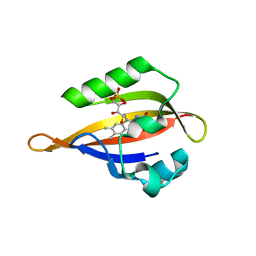 | | Crystal structure of the Phot-LOV1 domain from Chlamydomonas reinhardtii in illuminated state. Data set of a single crystal. | | Descriptor: | FLAVIN MONONUCLEOTIDE, putative blue light receptor | | Authors: | Fedorov, R, Schlichting, I, Hartmann, E, Domratcheva, T, Fuhrmann, M, Hegemann, P. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures and molecular mechanism of a light-induced signaling switch: The Phot-LOV1 domain from Chlamydomonas reinhardtii.
Biophys.J., 84, 2003
|
|
7LOE
 
 | | T4 lysozyme mutant L99A in complex with 1-fluoranylnaphthalene | | Descriptor: | 1-fluoranylnaphthalene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1K8Z
 
 | | CRYSTAL STRUCTURE OF THE TRYPTOPHAN SYNTHASE BETA-SER178PRO MUTANT COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID | | Descriptor: | N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-26 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the beta Ser178--> Pro mutant of tryptophan synthase. A "knock-out" allosteric enzyme.
J.Biol.Chem., 277, 2002
|
|
1Y7Y
 
 | | High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila | | Descriptor: | C.AhdI | | Authors: | McGeehan, J.E, Streeter, S.D, Papapanagiotou, I, Fox, G.C, Kneale, G.G. | | Deposit date: | 2004-12-10 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila.
J.Mol.Biol., 346, 2005
|
|
7LOF
 
 | | T4 lysozyme mutant L99A in complex with 2-butylthiophene | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-butylthiophene, BETA-MERCAPTOETHANOL, ... | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LOC
 
 | | T4 lysozyme mutant L99A in complex with 1-bromanyl-4-fluoranyl-benzene | | Descriptor: | 1-bromanyl-4-fluoranyl-benzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-02-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LOB
 
 | | T4 lysozyme mutant L99A in complex with 1-fluoro-2-[(prop-2-en-1-yl)oxy]benzene | | Descriptor: | 1-fluoro-2-[(prop-2-en-1-yl)oxy]benzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-02-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LX9
 
 | | T4 lysozyme mutant L99A | | Descriptor: | (but-3-en-1-yl)benzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Lysozyme | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-03-03 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LOJ
 
 | | T4 lysozyme mutant L99A in complex with 4-(3-phenylpropyl)aniline | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-(3-phenylpropyl)aniline, Lysozyme | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1NPK
 
 | | REFINED X-RAY STRUCTURE OF DICTYOSTELIUM NUCLEOSIDE DIPHOSPHATE KINASE AT 1,8 ANGSTROMS RESOLUTION | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Morera, S, Lebras, G, Lascu, I, Veron, M. | | Deposit date: | 1994-07-27 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined X-ray structure of Dictyostelium discoideum nucleoside diphosphate kinase at 1.8 A resolution.
J.Mol.Biol., 243, 1994
|
|
1K3O
 
 | | Crystal Structure Analysis of apo Glutathione S-Transferase | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1 | | Authors: | Le Trong, I, Stenkamp, R.E, Ibarra, C, Atkins, W.M, Adman, E.T. | | Deposit date: | 2001-10-03 | | Release date: | 2002-10-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.3-A resolution structure of human glutathione S-transferase with S-hexyl glutathione bound reveals possible extended ligandin binding site.
Proteins, 48, 2002
|
|
1K7F
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]VALINE ACID | | Descriptor: | N-[1H-INDOL-3-YL-ACETYL]VALINE ACID, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-19 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a new class of allosteric effectors complexed to tryptophan synthase.
J.Biol.Chem., 277, 2002
|
|
1K8X
 
 | | Crystal Structure Of AlphaT183V Mutant Of Tryptophan Synthase From Salmonella Typhimurium | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE, ... | | Authors: | Kulik, V, Weyand, M, Siedel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2001-10-26 | | Release date: | 2002-12-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | On the Role of AlphaThr183 in the Allosteric Regulation and Catalytic Mechanism of Tryptophan Synthase
J.Mol.Biol., 324, 2002
|
|
1K7X
 
 | | CRYSTAL STRUCTURE OF THE BETA-SER178PRO MUTANT OF TRYPTOPHAN SYNTHASE | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-22 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the beta Ser178--> Pro mutant of tryptophan synthase. A "knock-out" allosteric enzyme.
J.Biol.Chem., 277, 2002
|
|
1KA4
 
 | | Structure of Pyrococcus furiosus carboxypeptidase Nat-Pb | | Descriptor: | LEAD (II) ION, M32 carboxypeptidase | | Authors: | Arndt, J.W, Hao, B, Ramakrishnan, V, Cheng, T, Chan, S.I, Chan, M.K. | | Deposit date: | 2001-10-31 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a Novel Carboxypeptidase from the Hyperthermophilic Archaeon Pyrococcus furiosus
Structure, 10, 2002
|
|
1K3U
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]ASPARTIC ACID | | Descriptor: | N-[1H-INDOL-3-YL-ACETYL]ASPARTIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-04 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of a new class of allosteric effectors complexed to tryptophan synthase.
J.Biol.Chem., 277, 2002
|
|
1YBX
 
 | | Conserved hypothetical protein Cth-383 from Clostridium thermocellum | | Descriptor: | Conserved hypothetical protein, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Chang, J, Zhao, M, Habel, J, Kataeva, I, Xu, H, Chen, L, Lee, D, Nguyen, J, Chang, S.-H, Horanyi, P, Florence, Q, Zhou, W, Lin, D, Zhang, H, Ljundahl, L, Liu, Z.-J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conserved hypothetical protein Cth-383 from Clostridium thermocellum
To be published
|
|
1YC9
 
 | | The crystal structure of the outer membrane protein VceC from the bacterial pathogen Vibrio cholerae at 1.8 resolution | | Descriptor: | MERCURY (II) ION, multidrug resistance protein, octyl beta-D-glucopyranoside | | Authors: | Federici, L, Du, D, Walas, F, Matsumura, H, Fernandez-Recio, J, McKeegan, K.S, Borges-Walmsley, M.I, Luisi, B.F, Walmsley, A.R. | | Deposit date: | 2004-12-22 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the outer membrane protein VCEC from the bacterial pathogen vibrio cholerae at 1.8 A resolution
J.Biol.Chem., 280, 2005
|
|
1OLZ
 
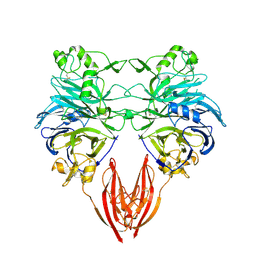 | | The ligand-binding face of the semaphorins revealed by the high resolution crystal structure of SEMA4D | | Descriptor: | SEMAPHORIN 4D | | Authors: | Love, C.A, Harlos, K, Mavaddat, N, Davis, S.J, Stuart, D.I, Jones, E.Y, Esnouf, R.M. | | Deposit date: | 2003-08-19 | | Release date: | 2003-09-11 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ligand-Binding Face of the Semaphorins Revealed by the High-Resolution Crystal Structure of Sema4D
Nat.Struct.Biol., 10, 2003
|
|
1K6H
 
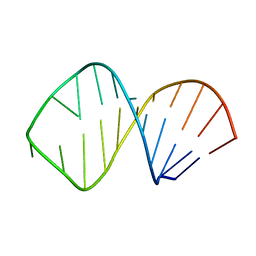 | | Solution Structure of Conserved AGNN Tetraloops: Insights into Rnt1p RNA processing | | Descriptor: | RNA (5'-R(P*GP*GP*CP*GP*UP*GP*UP*UP*CP*AP*GP*AP*AP*GP*AP*AP*CP*GP*CP*GP*CP*C)-3') | | Authors: | Lebars, I, Lamontagne, B, Yoshizawa, S, Abou Elela, S, Fourmy, D. | | Deposit date: | 2001-10-16 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of conserved AGNN tetraloops: insights into Rnt1p RNA processing.
EMBO J., 20, 2001
|
|
