2C1G
 
 | | Structure of Streptococcus pneumoniae peptidoglycan deacetylase (SpPgdA) | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, PEPTIDOGLYCAN GLCNAC DEACETYLASE, ... | | Authors: | Blair, D.E, Schuttelkopf, A.W, MacRae, J.I, van Aalten, D.M.F. | | Deposit date: | 2005-09-14 | | Release date: | 2005-09-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
6EQT
 
 | |
7KYU
 
 | | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1H-indole-5-carbonyl)oxy]-1H-benzotriazole, 3C-like proteinase | | Authors: | Tan, K, Maltseva, N.I, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate
To Be Published
|
|
6KB8
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW7647 co-crystals obtained by cross-seeding | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
3DRY
 
 | |
3RKV
 
 | | C-terminal domain of protein C56C10.10, a putative peptidylprolyl isomerase, from Caenorhabditis elegans | | Descriptor: | putative peptidylprolyl isomerase | | Authors: | Osipiuk, J, Tesar, C, Gu, M, Van Oosten-Hawle, P, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-18 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | C-terminal domain of protein C56C10.10, a putative peptidylprolyl isomerase, from Caenorhabditis elegans
To be Published
|
|
6TJE
 
 | |
1MW0
 
 | | Amylosucrase mutant E328Q co-crystallized with maltoheptaose then soaked with maltoheptaose. | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, amylosucrase, ... | | Authors: | Skov, L.K, Mirza, O, Sprogoe, D, Dar, I, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | Deposit date: | 2002-09-27 | | Release date: | 2002-12-18 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Oligosaccharide and Sucrose Complexes of Amylosucrase. STRUCTURAL IMPLICATIONS FOR THE POLYMERASE ACTIVITY
J.BIOL.CHEM., 277, 2002
|
|
7L63
 
 | |
6G7G
 
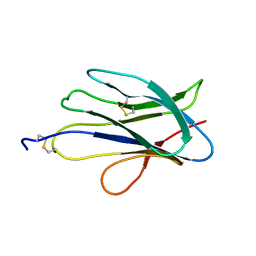 | | Structure of SPH (Self-Incompatibility Protein Homologue) proteins, a widespread family of small, highly stable, secreted proteins from plants | | Descriptor: | S-protein homolog 15 | | Authors: | Rajasekar, K.V, Coulthard, R.J, Ride, J.P, Ji, S, Winn, P.J, Wheeler, M.P, Hyde, E.I, Smith, L.J. | | Deposit date: | 2018-04-06 | | Release date: | 2019-03-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of SPH (self-incompatibility protein homologue) proteins: a widespread family of small, highly stable, secreted proteins.
Biochem.J., 476, 2019
|
|
6TE4
 
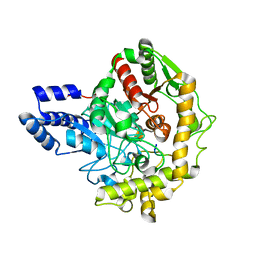 | | Structural insights into Pseudomonas aeruginosa Type six secretion system exported effector 8: Tse8 in complex with a peptide | | Descriptor: | Pro-Pro-Leu-Ala-Ser-Lys, Tse8 | | Authors: | Sainz-Polo, M.A, Capuni, R, Lucas, M, Altuna, J, Fucini, P, Montanchez, I, Albesa-Jove, D. | | Deposit date: | 2019-11-11 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural insights into Pseudomonas aeruginosaType six secretion system exported effector 8.
J.Struct.Biol., 212, 2020
|
|
3R2X
 
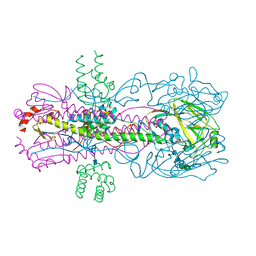 | |
1N1N
 
 | | Structure of Mispairing of the Deoxycytosine with Deoxyadenosine 5' to the 8,9-Dihydro-8-(N7-guanyl)-9-Hydroxy-Aflatoxin B1 Adduct | | Descriptor: | 5'-D(*AP*CP*AP*TP*CP*GP*AP*TP*CP*T)-3', 5'-D(*AP*GP*AP*TP*CP*AP*AP*TP*GP*T)-3', 8,9-DIHYDRO-9-HYDROXY-AFLATOXIN B1 | | Authors: | Stone, M.P, Giri, I. | | Deposit date: | 2002-10-18 | | Release date: | 2003-10-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Wobble dC.dA pairing 5' to the cationic guanine N7 8,9-dihydro-8-(N7-guanyl)-9-hydroxyaflatoxin B1 adduct: implications for nontargeted AFB1 mutagenesis.
Biochemistry, 42, 2003
|
|
6GF0
 
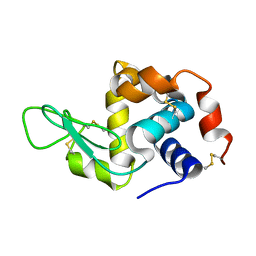 | | Lysozyme structure determined from SFX data using a Sheet-on-Sheet chipless chip | | Descriptor: | Lysozyme C | | Authors: | Doak, R.B, Gorel, A, Foucar, L, Gruenbein, M.L, Hilpert, M, Kloos, M, Nass Kovacs, G, Roome, C, Shoeman, R.L, Stricker, M, Tono, K, You, D, Ueda, K, Sherrel, D, Owen, R, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2018-04-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystallography on a chip - without the chip: sheet-on-sheet sandwich.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4ZET
 
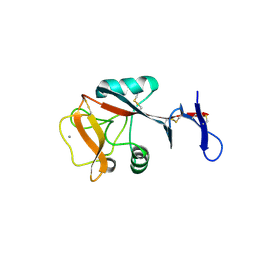 | | Blood dendritic cell antigen 2 (BDCA-2) complexed with GalGlcNAcMan | | Descriptor: | C-type lectin domain family 4 member C, CALCIUM ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Jegouzo, S.A.F, Feinberg, H, Dungarwalla, T, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2015-04-20 | | Release date: | 2015-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Novel Mechanism for Binding of Galactose-terminated Glycans by the C-type Carbohydrate Recognition Domain in Blood Dendritic Cell Antigen 2.
J.Biol.Chem., 290, 2015
|
|
1N6T
 
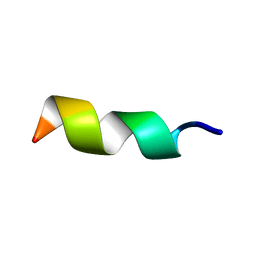 | |
1YT6
 
 | | NMR structure of peptide SD | | Descriptor: | peptide SD | | Authors: | Murata, T, Hemmi, H, Nakamura, S, Shimizu, K, Suzuki, Y, Yamaguchi, I. | | Deposit date: | 2005-02-10 | | Release date: | 2005-09-27 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure, epitope mapping, and docking simulation of a gibberellin mimic peptide as a peptidyl mimotope for a hydrophobic ligand.
Febs J., 272, 2005
|
|
3DRX
 
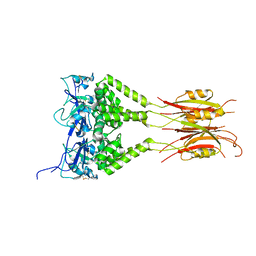 | |
1N7K
 
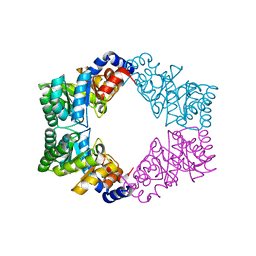 | | Unique tetrameric structure of deoxyribose phosphate aldolase from Aeropyrum pernix | | Descriptor: | deoxyribose-phosphate aldolase | | Authors: | Tsuge, H, Sakuraba, H, Shimoya, I, Katunuma, N, Ago, H, Miyano, M, Ohshima, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-15 | | Release date: | 2003-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The First Crystal Structure of Archaeal Aldolase. UNIQUE TETRAMERIC STRUCTURE of 2-DEOXY-D-RIBOSE-5-PHOSPHATE ALDOLASE FROM THE HYPERTHERMOPHILIC ARCHAEA Aeropyrum pernix.
J.Biol.Chem., 278, 2003
|
|
1N9N
 
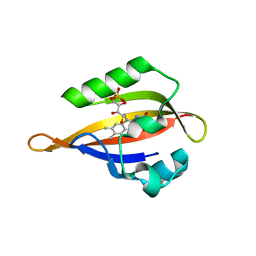 | | Crystal structure of the Phot-LOV1 domain from Chlamydomonas reinhardtii in illuminated state. Data set of a single crystal. | | Descriptor: | FLAVIN MONONUCLEOTIDE, putative blue light receptor | | Authors: | Fedorov, R, Schlichting, I, Hartmann, E, Domratcheva, T, Fuhrmann, M, Hegemann, P. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures and molecular mechanism of a light-induced signaling switch: The Phot-LOV1 domain from Chlamydomonas reinhardtii.
Biophys.J., 84, 2003
|
|
4N18
 
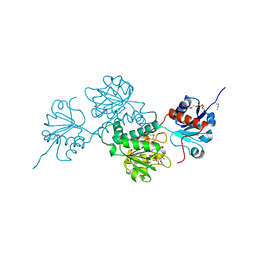 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase family protein from Klebsiella pneumoniae 342 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CITRIC ACID, D-isomer specific 2-hydroxyacid dehydrogenase family protein | | Authors: | Bacal, P, Shabalin, I.G, Cooper, D.R, Majorek, K.A, Osinski, T, Hillerich, B.S, Hammonds, J, Nawar, A, Stead, M, Chowdhury, S, Gizzi, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase family protein from Klebsiella pneumoniae 342
To be Published
|
|
1YFC
 
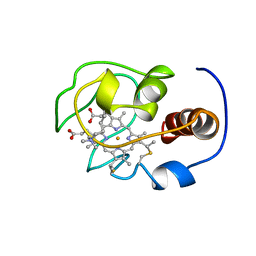 | | Solution nmr structure of a yeast iso-1-ferrocytochrome C | | Descriptor: | HEME C, YEAST ISO-1-FERROCYTOCHROME C | | Authors: | Baistrocchi, P, Banci, L, Bertini, I, Turano, P, Bren, K.L, Gray, H.B. | | Deposit date: | 1996-08-08 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Saccharomyces cerevisiae reduced iso-1-cytochrome c.
Biochemistry, 35, 1996
|
|
1YIC
 
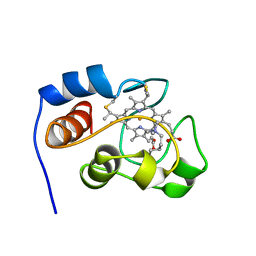 | | THE OXIDIZED SACCHAROMYCES CEREVISIAE ISO-1-CYTOCHROME C, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C, ISO-1, HEME C | | Authors: | Banci, L, Bertini, I, Bren, K.L, Gray, H.B, Sompornpisut, P, Turano, P. | | Deposit date: | 1997-02-18 | | Release date: | 1997-07-23 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of oxidized Saccharomyces cerevisiae iso-1-cytochrome c.
Biochemistry, 36, 1997
|
|
6GK5
 
 | |
6AZI
 
 | | 1.75 Angstrom Resolution Crystal Structure of D-alanyl-D-alanine Endopeptidase from Enterobacter cloacae in Complex with Covalently Bound Boronic Acid | | Descriptor: | BORATE ION, D-alanyl-D-alanine endopeptidase | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-11 | | Release date: | 2017-10-04 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom Resolution Crystal Structure of D-alanyl-D-alanine Endopeptidase from Enterobacter cloacae in Complex with Covalently Bound Boronic Acid.
To be Published
|
|
