8BGU
 
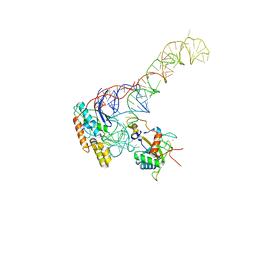 | | human MDM2-5S RNP | | Descriptor: | 5S rRNA, 60S ribosomal protein L11, 60S ribosomal protein L5, ... | | Authors: | Castillo, N, Thoms, M, Flemming, D, Hammaren, H.M, Buschauer, R, Ameismeier, M, Bassler, J, Beck, M, Beckmann, R, Hurt, E. | | Deposit date: | 2022-10-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of nascent 5S RNPs at the crossroad between ribosome assembly and MDM2-p53 pathways.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6ZQF
 
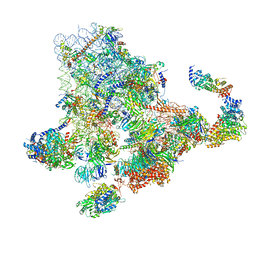 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-B (Poly-Ala) | | Descriptor: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
4V7F
 
 | | Arx1 pre-60S particle. | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Leidig, C, Thoms, M, Holdermann, I, Bradatsch, B, Berninghausen, O, Bange, G, Sinning, I, Hurt, E, Beckmann, R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle.
Nat Commun, 5, 2014
|
|
3M99
 
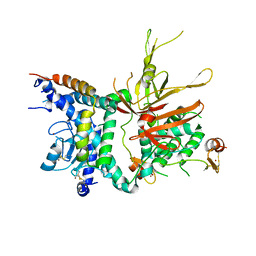 | | Structure of the Ubp8-Sgf11-Sgf73-Sus1 SAGA DUB module | | Descriptor: | Protein SUS1, SAGA-associated factor 11, SAGA-associated factor 73, ... | | Authors: | Kohler, A, Zimmerman, E, Schneider, M, Hurt, E, Zheng, N. | | Deposit date: | 2010-03-21 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module.
Cell(Cambridge,Mass.), 141, 2010
|
|
2MVF
 
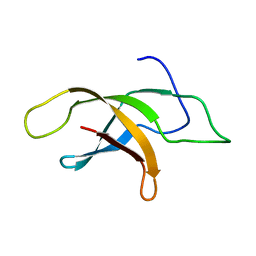 | | Structural insight into an essential assembly factor network on the pre-ribosome | | Descriptor: | Uncharacterized protein | | Authors: | Lee, W, Bassler, J, Paternoga, H, Holdermann, I, Thomas, M, Granneman, S, Barrio-Garcia, C, Nyarko, A, Stier, G, Clark, S.A, Schraivogel, D, Kallas, M, Beckmann, R, Tollervey, D, Barbar, E, Sinning, I, Hurt, E. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
9L5T
 
 | | Cryo-EM structure of the thermophile spliceosome (state B*Q2) | | Descriptor: | Anaphase-promoting complex subunit 4-like WD40 domain-containing protein, CCDC12, Delta(14)-sterol reductase, ... | | Authors: | Li, Y, Fischer, P, Wang, M, Yuan, R, Meng, W, Luehrmann, R, Lau, B, Hurt, E, Cheng, J. | | Deposit date: | 2024-12-23 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Cell Res., 35, 2025
|
|
9L5S
 
 | | Cryo-EM structure of the thermophile spliceosome (state B*Q1) | | Descriptor: | Anaphase-promoting complex subunit 4-like WD40 domain-containing protein, CCDC12, Delta(14)-sterol reductase, ... | | Authors: | Li, Y, Fischer, P, Wang, M, Yuan, R, Meng, W, Luehrmann, R, Lau, B, Hurt, E, Cheng, J. | | Deposit date: | 2024-12-23 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Cell Res., 35, 2025
|
|
9L5R
 
 | | Cryo-EM structure of the thermophile spliceosome (state ILS) | | Descriptor: | Anaphase-promoting complex subunit 4-like WD40 domain-containing protein, CCDC12, Cell cycle control protein, ... | | Authors: | Li, Y, Fischer, P, Wang, M, Yuan, R, Meng, W, Luehrmann, R, Lau, B, Hurt, E, Cheng, J. | | Deposit date: | 2024-12-23 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Cell Res., 35, 2025
|
|
7AJU
 
 | | Cryo-EM structure of the 90S-exosome super-complex (state Post-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
7AJT
 
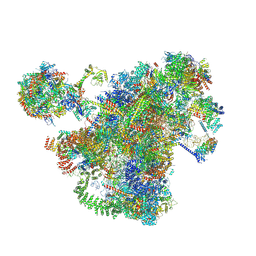 | | Cryo-EM structure of the 90S-exosome super-complex (state Pre-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
7OZS
 
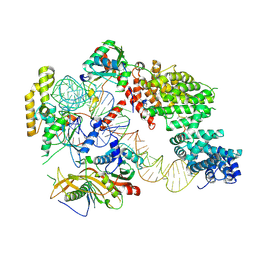 | | Structure of the hexameric 5S RNP from C. thermophilum | | Descriptor: | 5S rRNA, 60S ribosomal protein l5-like protein, Putative ribosomal protein, ... | | Authors: | Castillo, N, Thoms, M, Flemming, D, Hammaren, H.M, Buschauer, R, Ameismeier, M, Bassler, J, Beck, M, Beckmann, R, Hurt, E. | | Deposit date: | 2021-06-28 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of nascent 5S RNPs at the crossroad between ribosome assembly and MDM2-p53 pathways.
Nat.Struct.Mol.Biol., 2023
|
|
8QL1
 
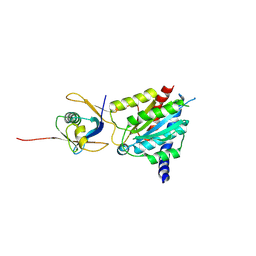 | | Crystal structure of the human MDN1-MIDAS/NLE1-UBL complex | | Descriptor: | MAGNESIUM ION, NDE1, Notchless protein homolog 1 | | Authors: | Wild, K, Fiorentino, F, Hurt, E, Sinning, I. | | Deposit date: | 2023-09-19 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Highly conserved ribosome biogenesis pathways between human and yeast revealed by the MDN1-NLE1 interaction and NLE1 containing pre-60S subunits.
Nucleic Acids Res., 53, 2025
|
|
3J2I
 
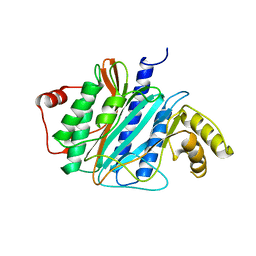 | | Structure of late pre-60S ribosomal subunits with nuclear export factor Arx1 bound at the peptide exit tunnel | | Descriptor: | Eukaryotic translation initiation factor 6, Proliferation-associated protein 2G4 | | Authors: | Bradatsch, B, Leidig, C, Granneman, S, Gnaedig, M, Tollervey, D, Boettcher, B, Beckmann, R, Hurt, E. | | Deposit date: | 2012-10-04 | | Release date: | 2012-11-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.9 Å) | | Cite: | Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1GO5
 
 | |
4GYG
 
 | | Crystal structure of the Rio2 kinase from Chaetomium thermophilum | | Descriptor: | Rio2 kinase | | Authors: | Ferreira-Cerca, S, Sagar, V, Schafer, T, Diop, M, Wesseling, A.M, Lu, H, Chai, E, Hurt, E, LaRonde-LeBlanc, N. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.482 Å) | | Cite: | ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4GYI
 
 | | Crystal structure of the Rio2 kinase-ADP/Mg2+-phosphoaspartate complex from Chaetomium thermophilum | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ferreira-Cerca, S, Sagar, V, Schafer, T, Diop, M, Wesseling, A.M, Lu, H, Chai, E, Hurt, E, LaRonde-LeBlanc, N. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4WJS
 
 | |
4WJV
 
 | | Crystal structure of Rsa4 in complex with the Nsa2 binding peptide | | Descriptor: | Maltose-binding periplasmic protein, Ribosome assembly protein 4, Ribosome biogenesis protein NSA2, ... | | Authors: | Holdermann, I, Paternoga, H, Bassler, J, Hurt, E, Sinning, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
4WJU
 
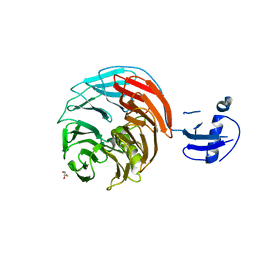 | | Crystal structure of Rsa4 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Ribosome assembly protein 4 | | Authors: | Holdermann, I, Bassler, J, Hurt, E, Sinning, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
6EML
 
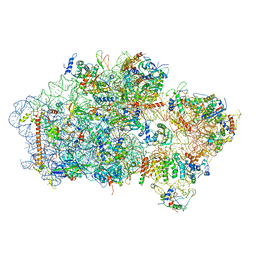 | | Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae | | Descriptor: | 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Heuer, A, Thomson, E, Schmidt, C, Berninghausen, O, Becker, T, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of a late pre-40S ribosomal subunit fromSaccharomyces cerevisiae.
Elife, 6, 2017
|
|
8I9P
 
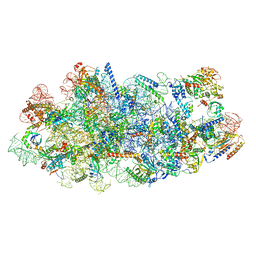 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State Mak16 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, 60S ribosomal protein L16-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
6QTA
 
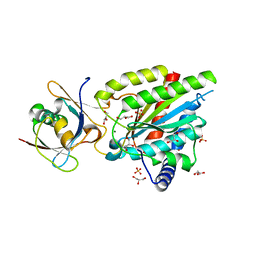 | | Crystal structure of Rea1-MIDAS/Rsa4-UBL complex from Chaetomium thermophilum | | Descriptor: | GLYCEROL, MAGNESIUM ION, Midasin,Midasin, ... | | Authors: | Ahmed, Y.L, Thoms, M, Hurt, E, Sinning, I. | | Deposit date: | 2019-02-22 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of Rea1-MIDAS bound to its ribosome assembly factor ligands resembling integrin-ligand-type complexes.
Nat Commun, 10, 2019
|
|
6QTB
 
 | | Crystal structure of Rea1-MIDAS/Ytm1-UBL complex from Chaetomium thermophilum | | Descriptor: | GLYCEROL, MAGNESIUM ION, Midasin,Midasin, ... | | Authors: | Ahmed, Y.L, Thoms, M, Hurt, E, Sinning, I. | | Deposit date: | 2019-02-22 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of Rea1-MIDAS bound to its ribosome assembly factor ligands resembling integrin-ligand-type complexes.
Nat Commun, 10, 2019
|
|
6QT8
 
 | | Crystal structure of Rea1-MIDAS domain from Chaetomium thermophilum | | Descriptor: | GLYCEROL, IODIDE ION, Midasin, ... | | Authors: | Ahmed, Y.L, Thoms, M, Hurt, E, Sinning, I. | | Deposit date: | 2019-02-22 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of Rea1-MIDAS bound to its ribosome assembly factor ligands resembling integrin-ligand-type complexes.
Nat Commun, 10, 2019
|
|
8I9X
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - Ytm1-1 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
