4FZS
 
 | | Structure of human SNX1 BAR domain | | Descriptor: | Sorting nexin-1 | | Authors: | van Weering, J.R.T, Sessions, R.B, Traer, C.J, Kloer, D.P, Bhatia, V.K, Stamou, D, Hurley, J.H, Cullen, P.J. | | Deposit date: | 2012-07-07 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular insight into vesicle-to-tubule membrane remodeling by SNX-BAR proteins
To be Published
|
|
2FIF
 
 | |
3R42
 
 | |
2ICP
 
 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 4.0. Northeast Structural Genomics Consortium TARGET ER390. | | Descriptor: | MAGNESIUM ION, antitoxin higa | | Authors: | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the bacterial antitoxin HigA from Escherichia coli.
To be Published
|
|
4IKN
 
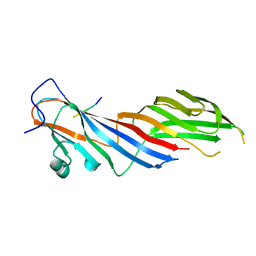 | | Crystal structure of adaptor protein complex 3 (AP-3) mu3A subunit C-terminal domain, in complex with a sorting peptide from TGN38 | | Descriptor: | AP-3 complex subunit mu-1, Trans-Golgi network integral membrane protein TGN38 | | Authors: | Mardones, G.A, Kloer, D.P, Burgos, P.V, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2012-12-26 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for the recognition of tyrosine-based sorting signals by the mu 3A subunit of the AP-3 adaptor complex.
J.Biol.Chem., 288, 2013
|
|
2P22
 
 | | Structure of the Yeast ESCRT-I Heterotetramer Core | | Descriptor: | Hypothetical 12.0 kDa protein in ADE3-SER2 intergenic region, Protein SRN2, SULFATE ION, ... | | Authors: | Kostelansky, M.S, Hurley, J.H. | | Deposit date: | 2007-03-06 | | Release date: | 2007-06-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer.
Cell(Cambridge,Mass.), 129, 2007
|
|
2FID
 
 | |
4HMY
 
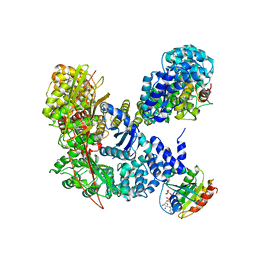 | | Structural basis for recruitment and activation of the AP-1 clathrin adaptor complex by Arf1 | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Ren, X, Farias, G.G, Canagarajah, B.J, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2012-10-18 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Structural Basis for Recruitment and Activation of the AP-1 Clathrin Adaptor Complex by Arf1.
Cell(Cambridge,Mass.), 152, 2013
|
|
2ICT
 
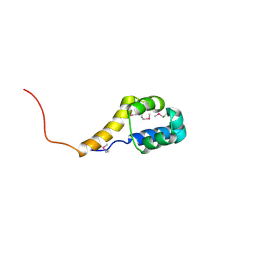 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 8.5. Northeast Structural Genomics TARGET ER390. | | Descriptor: | antitoxin higa | | Authors: | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
2FAU
 
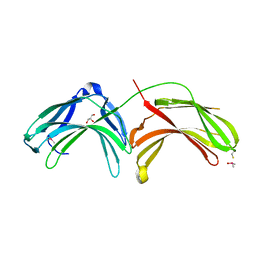 | | Crystal structure of human vps26 | | Descriptor: | GLYCEROL, Vacuolar protein sorting 26 | | Authors: | Shi, H, Rojas, R, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2005-12-08 | | Release date: | 2006-05-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its C-terminal domain.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1QAT
 
 | |
1QAS
 
 | |
4HPQ
 
 | | Crystal Structure of the Atg17-Atg31-Atg29 Complex | | Descriptor: | Atg17, Atg29, Atg31 | | Authors: | Stanley, R.E, Ragusa, M.J, Hurley, J.H. | | Deposit date: | 2012-10-24 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Architecture of the atg17 complex as a scaffold for autophagosome biogenesis.
Cell(Cambridge,Mass.), 151, 2012
|
|
3PFQ
 
 | | Crystal Structure and Allosteric Activation of Protein Kinase C beta II | | Descriptor: | CALCIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein kinase C beta type, ... | | Authors: | Leonard, T.A, Rozycki, B, Saidi, L.F, Hummer, G, Hurley, J.H. | | Deposit date: | 2010-10-28 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure and Allosteric Activation of Protein Kinase C beta II
Cell(Cambridge,Mass.), 144, 2011
|
|
4K7H
 
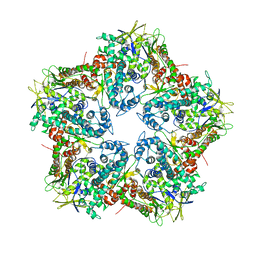 | | Major capsid protein P1 of the Pseudomonas phage phi6 | | Descriptor: | Major inner protein P1 | | Authors: | Boura, E, Nemecek, D, Plevka, P, Steven, C.A, Hurley, J.H. | | Deposit date: | 2013-04-17 | | Release date: | 2013-08-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5964 Å) | | Cite: | Subunit Folds and Maturation Pathway of a dsRNA Virus Capsid.
Structure, 21, 2013
|
|
4J2G
 
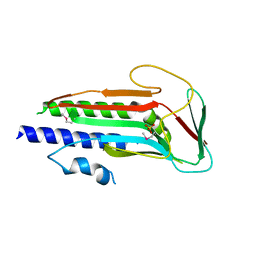 | | Atg13 HORMA domain | | Descriptor: | KLTH0A00704p, SULFATE ION | | Authors: | Jao, C, Stanley, R.E, Ragusa, M.J, Hurley, J.H. | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-20 | | Last modified: | 2013-04-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A HORMA domain in Atg13 mediates PI 3-kinase recruitment in autophagy.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1ZI7
 
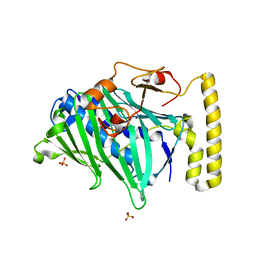 | | Structure of truncated yeast oxysterol binding protein Osh4 | | Descriptor: | KES1 protein, SULFATE ION | | Authors: | Im, Y.J, Raychaudhuri, S, Prinz, W.A, Hurley, J.H. | | Deposit date: | 2005-04-27 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural mechanism for sterol sensing and transport by OSBP-related proteins
Nature, 437, 2005
|
|
2OJQ
 
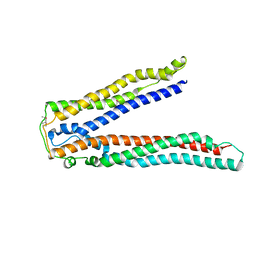 | | Crystal structure of Alix V domain | | Descriptor: | Programmed cell death 6-interacting protein | | Authors: | Lee, S, Hurley, J.H. | | Deposit date: | 2007-01-13 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural basis for viral late-domain binding to Alix
Nat.Struct.Mol.Biol., 14, 2007
|
|
1XA6
 
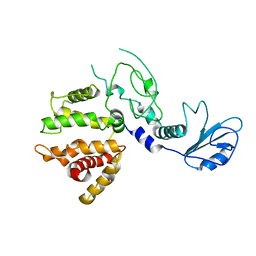 | | Crystal Structure of the Human Beta2-Chimaerin | | Descriptor: | Beta2-chimaerin, ZINC ION | | Authors: | Canagarajah, B, Leskow, F.C, Ho, J.Y, Mischak, H, Saidi, L.F, Kazanietz, M.G, Hurley, J.H. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mechanism for lipid activation of the Rac-specific GAP, beta2-chimaerin.
Cell(Cambridge,Mass.), 119, 2004
|
|
1XHB
 
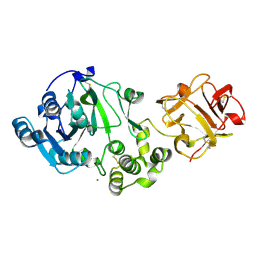 | | The Crystal Structure of UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase-T1 | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Polypeptide N-acetylgalactosaminyltransferase 1, ... | | Authors: | Fritz, T.A, Hurley, J.H, Trinh, L.B, Shiloach, J, Tabak, L.A. | | Deposit date: | 2004-09-17 | | Release date: | 2004-10-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The beginnings of mucin biosynthesis: The crystal structure of UDP-GalNAc:polypeptide {alpha}-N-acetylgalactosaminyltransferase-T1
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1YD8
 
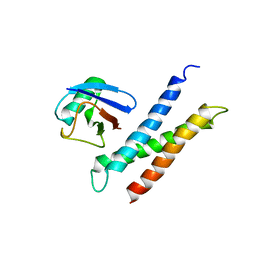 | | COMPLEX OF HUMAN GGA3 GAT DOMAIN AND UBIQUITIN | | Descriptor: | ADP-RIBOSYLATION FACTOR BINDING PROTEIN GGA3, UBIQUIN | | Authors: | Prag, G, Lee, S, Mattera, R, Arighi, C.N, Beach, B.M, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2004-12-23 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanism for ubiquitinated-cargo recognition by the Golgi-localized, {gamma}-ear-containing, ADP-ribosylation-factor-binding proteins
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1ZHX
 
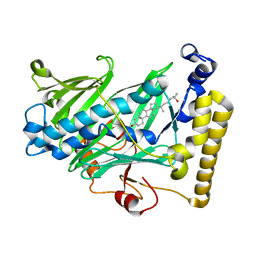 | | Structure of yeast oxysterol binding protein Osh4 in complex with 25-hydroxycholesterol | | Descriptor: | 25-HYDROXYCHOLESTEROL, KES1 protein | | Authors: | Im, Y.J, Raychaudhuri, S, Prinz, W.A, Hurley, J.H. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural mechanism for sterol sensing and transport by OSBP-related proteins
Nature, 437, 2005
|
|
1ZHW
 
 | | Structure of yeast oxysterol binding protein Osh4 in complex with 20-hydroxycholesterol | | Descriptor: | 20-HYDROXYCHOLESTEROL, KES1 protein, LEAD (II) ION | | Authors: | Im, Y.J, Raychaudhuri, S, Prinz, W.A, Hurley, J.H. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural mechanism for sterol sensing and transport by OSBP-related proteins
Nature, 437, 2005
|
|
1Z2N
 
 | | Inositol 1,3,4-trisphosphate 5/6-kinase complexed Mg2+/ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, inositol 1,3,4-trisphosphate 5/6-kinase | | Authors: | Miller, G.J, Wilson, M.P, Majerus, P.W, Hurley, J.H. | | Deposit date: | 2005-03-08 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase.
Mol.Cell, 18, 2005
|
|
1Z2P
 
 | | Inositol 1,3,4-trisphosphate 5/6-Kinase in complex with Mg2+/AMP-PCP/Ins(1,3,4)P3 | | Descriptor: | (1S,3S,4S)-1,3,4-TRIPHOSPHO-MYO-INOSITOL, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Miller, G.J, Wilson, M.P, Majerus, P.W, Hurley, J.H. | | Deposit date: | 2005-03-08 | | Release date: | 2005-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase.
Mol.Cell, 18, 2005
|
|
