1XVW
 
 | | Crystal Structure of AhpE from Mycobacterium tuberculosis, a 1-Cys peroxiredoxin | | Descriptor: | Hypothetical protein Rv2238c/MT2298 | | Authors: | Li, S, Peterson, N.A, Kim, M.Y, Kim, C.Y, Hung, L.W, Yu, M, Lekin, T, Segelke, B.W, Lott, J.S, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-10-28 | | Release date: | 2005-02-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of AhpE from Mycobacterium tuberculosis, a 1-Cys Peroxiredoxin
J.Mol.Biol., 346, 2005
|
|
6O7F
 
 | |
3LP6
 
 | | Crystal Structure of Rv3275c-E60A from Mycobacterium tuberculosis at 1.7A resolution | | Descriptor: | FORMIC ACID, GLYCEROL, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Kim, H, Yu, M, Hung, L.-W, Terwilliger, T.C, Kim, C.-Y, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Crystal Structure of Rv3275c-E60A from Mycobacterium tuberculosis at 1.7A
Resolution
To be Published
|
|
1SBZ
 
 | | Crystal Structure of dodecameric FMN-dependent Ubix-like Decarboxylase from Escherichia coli O157:H7 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Probable aromatic acid decarboxylase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Tocilj, A, Hung, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-02-11 | | Release date: | 2004-10-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a dodecameric FMN-dependent UbiX-like decarboxylase (Pad1) from Escherichia coli O157: H7.
Protein Sci., 13, 2004
|
|
4KF4
 
 | | Crystal Structure of sfCherry | | Descriptor: | fluorescent protein sfCherry | | Authors: | Nguyen, H.B, Hung, L.-W, Yeates, T.O, Waldo, G.S, Terwilliger, T.C. | | Deposit date: | 2013-04-26 | | Release date: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Split green fluorescent protein as a modular binding partner for protein crystallization.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3FHK
 
 | | Crystal structure of APC1446, B.subtilis YphP disulfide isomerase | | Descriptor: | SULFATE ION, UPF0403 protein yphP | | Authors: | Derewenda, U, Boczek, T, Cooper, D.R, Yu, M, Hung, L, Derewenda, Z.S, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2008-12-09 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of Bacillus subtilis YphP, a prokaryotic disulfide isomerase with a CXC catalytic motif .
Biochemistry, 48, 2009
|
|
3H1J
 
 | | Stigmatellin-bound cytochrome bc1 complex from chicken | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, Coenzyme Q10, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.-I, Kim, K.K, Hung, L.-W, Crofts, A.R, Berry, E.A, Kim, S.-H. | | Deposit date: | 2009-04-12 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
4P0M
 
 | | Crystal structure of an evolved putative penicillin-binding protein homolog, Rv2911, from Mycobacterium tuberculosis | | Descriptor: | D-alanyl-D-alanine carboxypeptidase | | Authors: | Krieger, I, Yu, M, Bursey, E, Hung, L.-W, Terwilliger, T.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subfamily-Specific Adaptations in the Structures of Two Penicillin-Binding Proteins from Mycobacterium tuberculosis.
Plos One, 9, 2014
|
|
7WLV
 
 | | Crystal Structure of the Multidrug effulx transporter BpeF from Burkholderia pseudomallei. | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Efflux pump membrane transporter | | Authors: | Kato, T, Hung, L.-W, Yamashita, E, Okada, U, Terwilliger, T.C, Murakami, S. | | Deposit date: | 2022-01-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of multidrug efflux transporters from Burkholderia pseudomallei suggest details of transport mechanism.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1XXU
 
 | | Crystal Structure of AhpE from Mycrobacterium tuberculosis, a 1-Cys peroxiredoxin | | Descriptor: | Hypothetical protein Rv2238c/MT2298 | | Authors: | Li, S, Peterson, N.A, Kim, M.Y, Kim, C.Y, Hung, L.W, Yu, M, Lekin, T, Segelke, B.W, Lott, J.S, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-11-08 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of AhpE from Mycobacterium tuberculosis, a 1-Cys Peroxiredoxin
J.Mol.Biol., 346, 2005
|
|
2A2J
 
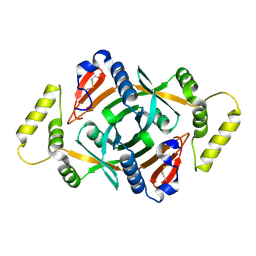 | | Crystal structure of a putative pyridoxine 5'-phosphate oxidase (Rv2607) from Mycobacterium tuberculosis | | Descriptor: | Pyridoxamine 5'-phosphate oxidase | | Authors: | Pedelacq, J.-D, Rho, B.-S, Kim, C.-Y, Waldo, G.S, Lekin, T.P, Segelke, B.W, Rupp, B, Hung, L.-W, Kim, S.-I, Terwilliger, T.C, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2005-06-22 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative pyridoxine 5'-phosphate oxidase (Rv2607) from Mycobacterium tuberculosis.
Proteins, 62, 2005
|
|
3U3Q
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2013-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U3P
 
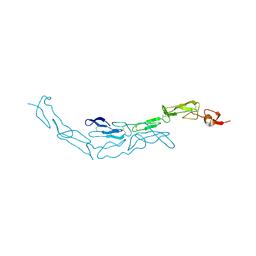 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2013-07-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U3V
 
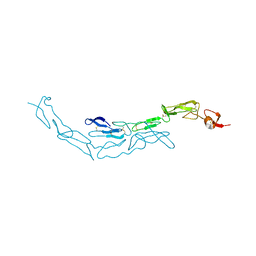 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4KF5
 
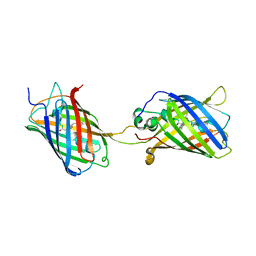 | | Crystal Structure of Split GFP complexed with engineered sfCherry with an insertion of GFP fragment | | Descriptor: | fluorescent protein GFP1-9, fluorescent protein sfCherry+GFP10-11 | | Authors: | Nguyen, H.B, Hung, L.-W, Yeates, T.O, Waldo, G.S, Terwilliger, T.C. | | Deposit date: | 2013-04-26 | | Release date: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Split green fluorescent protein as a modular binding partner for protein crystallization.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3U3T
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U3S
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2013-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1MJH
 
 | | Structure-based assignment of the biochemical function of hypothetical protein MJ0577: A test case of structural genomics | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PROTEIN (ATP-BINDING DOMAIN OF PROTEIN MJ0577) | | Authors: | Zarembinski, T.I, Hung, L.-W, Mueller-Dieckmann, H.J, Kim, K.-K, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 1998-11-04 | | Release date: | 1998-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based assignment of the biochemical function of a hypothetical protein: a test case of structural genomics.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4JBJ
 
 | | Structural mimicry for functional antagonism | | Descriptor: | Interferon-activable protein 202 | | Authors: | Ru, H, Ni, X, Ma, F, Zhao, L, Ding, W, Hung, L.-W, Shaw, N, Cheng, G, Liu, Z.-J. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Structural basis for termination of AIM2-mediated signaling by p202
Cell Res., 23, 2013
|
|
4JBM
 
 | | Structure of murine DNA binding protein bound with ds DNA | | Descriptor: | DNA (5'-D(*GP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), Interferon-inducible protein AIM2 | | Authors: | Ru, H, Ni, X, Crowley, C, Zhao, L, Ding, W, Hung, L.-W, Shaw, N, Cheng, G, Liu, Z.-J. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | Structural basis for termination of AIM2-mediated signaling by p202
Cell Res., 23, 2013
|
|
4JBK
 
 | | Molecular basis for abrogation of activation of pro-inflammatory cytokines | | Descriptor: | DNA (5'-D(P*GP*GP*AP*AP*TP*TP*AP*TP*AP*AP*TP*TP*CP*C)-3'), Interferon-activable protein 202 | | Authors: | Ru, H, Ni, X, Crowley, C, Zhao, L, Ding, W, Hung, L.-W, Shaw, N, Cheng, G, Liu, Z.-J. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.963 Å) | | Cite: | Structural basis for termination of AIM2-mediated signaling by p202
Cell Res., 23, 2013
|
|
7WLS
 
 | | Crystal structure of the multidrug efflux transporter BpeB from Burkholderia pseudomallei | | Descriptor: | Efflux pump membrane transporter, TETRAETHYLENE GLYCOL, UNDECYL-MALTOSIDE | | Authors: | Kato, T, Hung, L.-W, Yamashita, E, Okada, U, Terwilliger, T.C, Murakami, S. | | Deposit date: | 2022-01-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structures of multidrug efflux transporters from Burkholderia pseudomallei suggest details of transport mechanism.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4LMP
 
 | | Mycobacterium tuberculosis L-alanine dehydrogenase x-ray structure in complex with N6-methyl adenosine | | Descriptor: | Alanine dehydrogenase, GLYCEROL, N-methyladenosine, ... | | Authors: | Kim, H.-B, Hung, L.-W, Goulding, C.W, Terwilliger, T.C, Kim, C.-Y, Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-07-10 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Drug target analysis by dye-ligand affinity chromatography
To be Published
|
|
3H1H
 
 | | Cytochrome bc1 complex from chicken | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, CYTOCHROME C1, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.I, Kim, K.K, Hung, L.W, Crofts, A.R, Berry, E.A, Kim, S.H. | | Deposit date: | 2009-04-12 | | Release date: | 2009-04-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
3H1I
 
 | | Stigmatellin and antimycin bound cytochrome bc1 complex from chicken | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-METHYL-BUTYRIC ACID 3-(3-FORMYLAMINO-2-HYDROXY-BENZOYLAMINO)-8-HEPTYL-2,6-DIMETHYL-4,9-DIOXO-[1,5]DIOXONAN-7-YL ESTER, CARDIOLIPIN, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.I, Kim, K.K, Hung, L.W, Crofts, A.R, Berry, E.A, Kim, S.H. | | Deposit date: | 2009-04-12 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
