6OKB
 
 | | Prohead 2 of the phage T5 | | Descriptor: | Major capsid protein | | Authors: | Huet, A, Duda, R.L, Boulanger, P, Conway, J.F. | | Deposit date: | 2019-04-12 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Capsid expansion of bacteriophage T5 revealed by high resolution cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OMA
 
 | |
6OMC
 
 | | capsid of T5 virion | | Descriptor: | Major capsid protein | | Authors: | Huet, A, Duda, R.L, Boulanger, P, Conway, J.F. | | Deposit date: | 2019-04-18 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Capsid expansion of bacteriophage T5 revealed by high resolution cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8FQL
 
 | | Portal vertex of HK97 phage | | Descriptor: | Portal protein | | Authors: | Huet, A, Oh, B, Maurer, J, Duda, R.L, Conway, J.F. | | Deposit date: | 2023-01-06 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A symmetry mismatch unraveled: How phage HK97 scaffold flexibly accommodates a 12-fold pore at a 5-fold viral capsid vertex.
Sci Adv, 9, 2023
|
|
8FQK
 
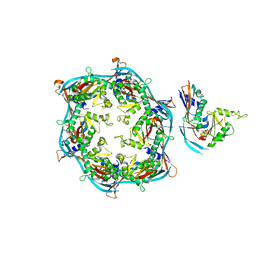 | | Asymmetric unit of HK97 phage prohead I | | Descriptor: | Scaffolding domain delta | | Authors: | Huet, A, Oh, B, Maurer, J, Duda, R.L, Conway, J.F. | | Deposit date: | 2023-01-06 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A symmetry mismatch unraveled: How phage HK97 scaffold flexibly accommodates a 12-fold pore at a 5-fold viral capsid vertex.
Sci Adv, 9, 2023
|
|
8OMI
 
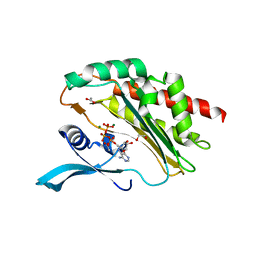 | | Crystal structure of the inositol hexakisphosphate kinase EhIP6KA M85 variant in complex with ATP and Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schuetz, A, Aguirre, T, Fiedler, D. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | An unconventional gatekeeper mutation sensitizes inositol hexakisphosphate kinases to an allosteric inhibitor.
Elife, 12, 2023
|
|
8CJL
 
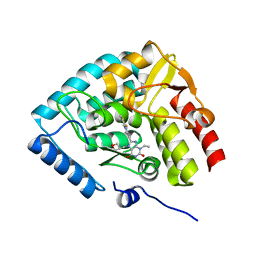 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor TPT-004 | | Descriptor: | 3-ethyl-8-[(2-methyl-5~{H}-imidazo[2,1-b][1,3]thiazol-6-yl)methyl]-7-(oxetan-3-ylmethyl)purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8CJO
 
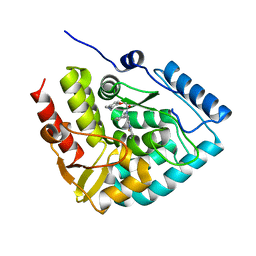 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-06-004 | | Descriptor: | 1-[(3~{S})-3-(4-chloranyl-2-fluoranyl-phenyl)-1,4,8-triazatricyclo[7.4.0.0^{2,7}]trideca-2(7),8-dien-4-yl]-2-(2-ethyl-6-methyl-pyridin-3-yl)oxy-ethanone, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86633706 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8CJM
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-07-047 | | Descriptor: | 7-(cyclobutylmethyl)-3-ethyl-8-(5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-2-ylmethyl)purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8CJI
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-07-052 | | Descriptor: | FE (III) ION, Tryptophan 5-hydroxylase 1, methyl (2~{S})-2-azanyl-3-[[3-[[3-ethyl-2,6-bis(oxidanylidene)-8-(5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-2-ylmethyl)purin-7-yl]methyl]phenyl]carbonylamino]propanoate | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8CJN
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-06-070 | | Descriptor: | 3-ethyl-7-[(4-phenylphenyl)methyl]-8-(5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-2-ylmethyl)purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68080938 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
4TXE
 
 | | ScCTS1 in complex with compound 5 | | Descriptor: | (2S)-1-(2,3-dihydro-1H-inden-2-ylamino)-3-(3,4-dimethylphenoxy)propan-2-ol, Endochitinase | | Authors: | Schuettelkopf, A.W, van Aalten, D.M.F. | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Screening-based discovery of Aspergillus fumigatus plant-type chitinase inhibitors
FEBS Lett, 588, 2014
|
|
8CJJ
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-06-057 | | Descriptor: | 3-ethyl-7-(phenylmethyl)-8-(5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-2-ylmethyl)purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66415656 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8CJK
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-06-098 | | Descriptor: | 3-ethyl-8-[(2-methylimidazo[2,1-b][1,3]thiazol-6-yl)methyl]-7-[[4-(1-methylpyrazol-3-yl)phenyl]methyl]purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45914972 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
4ULW
 
 | |
6EKU
 
 | | Vibrio cholerae neuraminidase complexed with zanamivir | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schuetz, A, Heinemann, U. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Different Inhibitory Potencies of Oseltamivir Carboxylate, Zanamivir, and Several Tannins on Bacterial and Viral Neuraminidases as Assessed in a Cell-Free Fluorescence-Based Enzyme Inhibition Assay.
Molecules, 22, 2017
|
|
3TT9
 
 | | Crystal structure of the stable degradation fragment of human plakophilin 2 isoform a (PKP2a) C752R variant | | Descriptor: | GLYCEROL, Plakophilin-2 | | Authors: | Schuetz, A, Roske, Y, Gerull, B, Heinemann, U. | | Deposit date: | 2011-09-14 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular insights into arrhythmogenic right ventricular cardiomyopathy caused by plakophilin-2 missense mutations.
Circ Cardiovasc Genet, 5, 2012
|
|
1E92
 
 | | Pteridine reductase 1 from Leishmania major complexed with NADP+ and dihydrobiopterin | | Descriptor: | 1,2-ETHANEDIOL, 7,8-DIHYDROBIOPTERIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schuettelkopf, A.W, Hunter, W.N. | | Deposit date: | 2000-10-04 | | Release date: | 2001-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pteridine Reductase Mechanism Correlates Pterin Metabolism with Drug Resistance in Trypanosomatid Parasites
Nat.Struct.Biol., 8, 2001
|
|
6EKS
 
 | | Vibrio cholerae neuraminidase complexed with oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Schuetz, A, Heinemann, U. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Different Inhibitory Potencies of Oseltamivir Carboxylate, Zanamivir, and Several Tannins on Bacterial and Viral Neuraminidases as Assessed in a Cell-Free Fluorescence-Based Enzyme Inhibition Assay.
Molecules, 22, 2017
|
|
2CH9
 
 | | Crystal structure of dimeric human cystatin F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Schuettelkopf, A.W, van Aalten, D.M.F. | | Deposit date: | 2006-03-13 | | Release date: | 2006-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Reduction-Dependent Activation of Human Cystatin F.
J.Biol.Chem., 281, 2006
|
|
4UAL
 
 | | MRCK beta in complex with BDP00005290 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-1-(piperidin-4-yl)-N-[3-(pyridin-2-yl)-1H-pyrazol-4-yl]-1H-pyrazole-3-carboxamide, CHLORIDE ION, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2014-08-10 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A novel small-molecule MRCK inhibitor blocks cancer cell invasion.
Cell Commun. Signal, 12, 2014
|
|
7ZIH
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor AG-01-128 | | Descriptor: | 8-(1~{H}-benzimidazol-2-ylmethyl)-3-ethyl-7-(phenylmethyl)purine-2,6-dione, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Schuetz, A, Gogolin, A, Pfeifer, J, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46890831 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
4UAK
 
 | | MRCK beta in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2014-08-10 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A novel small-molecule MRCK inhibitor blocks cancer cell invasion.
Cell Commun. Signal, 12, 2014
|
|
7ZII
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-05-193 | | Descriptor: | 8-(5~{H}-[1,3]dioxolo[4,5-f]benzimidazol-6-ylmethyl)-7-(phenylmethyl)-3-propyl-purine-2,6-dione, FE (III) ION, GLYCEROL, ... | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6280005 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIJ
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-05-080 | | Descriptor: | 8-(1~{H}-benzimidazol-2-ylmethyl)-3-cyclopropyl-7-(phenylmethyl)purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94678366 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
