1HH2
 
 | | Crystal structure of NusA from Thermotoga maritima | | Descriptor: | N UTILIZATION SUBSTANCE PROTEIN A | | Authors: | Worbs, M, Bourenkov, G.P, Bartunik, H.D, Huber, R, Wahl, M.C. | | Deposit date: | 2000-12-18 | | Release date: | 2001-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Extended RNA Binding Surface Through Arrayed S1 and Kh Domains in Transcription Factor Nusa
Mol.Cell, 7, 2001
|
|
1N08
 
 | | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ZINC ION, putative riboflavin kinase | | Authors: | Bauer, S, Kemter, K, Bacher, A, Huber, R, Fischer, M, Steinbacher, S. | | Deposit date: | 2002-10-11 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold
J.Mol.Biol., 326, 2003
|
|
1N31
 
 | | Structure of A Catalytically Inactive Mutant (K223A) of C-DES with a Substrate (Cystine) Linked to the Co-Factor | | Descriptor: | CYSTEINE, L-cysteine/cystine lyase C-DES, POTASSIUM ION, ... | | Authors: | Kaiser, J.T, Bruno, S, Clausen, T, Huber, R, Schiaretti, F, Mozzarelli, A, Kessler, D. | | Deposit date: | 2002-10-25 | | Release date: | 2003-01-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Snapshots of the Cystine Lyase "C-DES" during Catalysis: Studies in Solution and in the Crystalline State
J.Biol.Chem., 278, 2003
|
|
1MT3
 
 | | Crystal Structure of the Tricorn Interacting Factor Selenomethionine-F1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Proline iminopeptidase | | Authors: | Goettig, P, Groll, M, Kim, J.-S, Huber, R, Brandstetter, H. | | Deposit date: | 2002-09-20 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism
Embo J., 21, 2002
|
|
1HZ4
 
 | | CRYSTAL STRUCTURE OF TRANSCRIPTION FACTOR MALT DOMAIN III | | Descriptor: | BENZOIC ACID, GLYCEROL, MALT REGULATORY PROTEIN, ... | | Authors: | Steegborn, C, Danot, O, Clausen, T, Huber, R. | | Deposit date: | 2001-01-23 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of transcription factor MalT domain III: a novel helix repeat fold implicated in regulated oligomerization.
Structure, 9, 2001
|
|
1SEP
 
 | | MOUSE SEPIAPTERIN REDUCTASE COMPLEXED WITH NADP AND SEPIAPTERIN | | Descriptor: | BIOPTERIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SEPIAPTERIN REDUCTASE | | Authors: | Auerbach, G, Herrmann, A, Guetlich, M, Fischer, M, Jacob, U, Bacher, A, Huber, R. | | Deposit date: | 1997-05-23 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The 1.25 A crystal structure of sepiapterin reductase reveals its binding mode to pterins and brain neurotransmitters.
EMBO J., 16, 1997
|
|
1N07
 
 | | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, PUTATIVE riboflavin kinase | | Authors: | Bauer, S, Kemter, K, Bacher, A, Huber, R, Fischer, M, Steinbacher, S. | | Deposit date: | 2002-10-11 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold
J.Mol.Biol., 326, 2003
|
|
1CZV
 
 | | CRYSTAL STRUCTURE OF THE C2 DOMAIN OF HUMAN COAGULATION FACTOR V: DIMERIC CRYSTAL FORM | | Descriptor: | PROTEIN (COAGULATION FACTOR V) | | Authors: | Macedo-Ribeiro, S, Bode, W, Huber, R, Kane, W.H, Fuentes-Prior, P. | | Deposit date: | 1999-09-07 | | Release date: | 1999-11-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the membrane-binding C2 domain of human coagulation factor V.
Nature, 402, 1999
|
|
1CJA
 
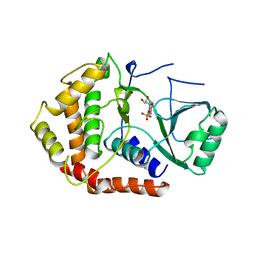 | | ACTIN-FRAGMIN KINASE, CATALYTIC DOMAIN FROM PHYSARUM POLYCEPHALUM | | Descriptor: | ADENOSINE MONOPHOSPHATE, PROTEIN (ACTIN-FRAGMIN KINASE) | | Authors: | Steinbacher, S, Hof, P, Eichinger, L, Schleicher, M, Gettemans, J, Vandekerckhove, J, Huber, R, Benz, J. | | Deposit date: | 1999-04-08 | | Release date: | 1999-06-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of the Physarum polycephalum actin-fragmin kinase: an atypical protein kinase with a specialized substrate-binding domain.
EMBO J., 18, 1999
|
|
1ICQ
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO COMPLEXED WITH 9R,13R-OPDA | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, 9R,13R-12-OXOPHYTODIENOIC ACID, FLAVIN MONONUCLEOTIDE | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1CZT
 
 | | CRYSTAL STRUCTURE OF THE C2 DOMAIN OF HUMAN COAGULATION FACTOR V | | Descriptor: | PROTEIN (COAGULATION FACTOR V) | | Authors: | Macedo-Ribeiro, S, Bode, W, Huber, R, Kane, W.H, Fuentes-Prior, P. | | Deposit date: | 1999-09-07 | | Release date: | 1999-11-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of the membrane-binding C2 domain of human coagulation factor V.
Nature, 402, 1999
|
|
1CZS
 
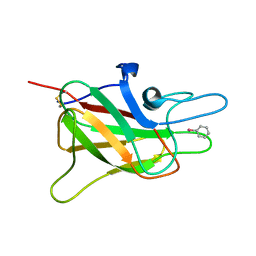 | | CRYSTAL STRUCTURE OF THE C2 DOMAIN OF HUMAN COAGULATION FACTOR V: COMPLEX WITH PHENYLMERCURY | | Descriptor: | PHENYLMERCURY, PROTEIN (COAGULATION FACTOR V) | | Authors: | Macedo-Ribeiro, S, Bode, W, Huber, R, Kane, W.H, Fuentes-Prior, P. | | Deposit date: | 1999-09-07 | | Release date: | 1999-11-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the membrane-binding C2 domain of human coagulation factor V.
Nature, 402, 1999
|
|
1N8P
 
 | | Crystal Structure of cystathionine gamma-lyase from yeast | | Descriptor: | Cystathionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Messerschmidt, A, Worbs, M, Steegborn, C, Wahl, M.C, Huber, R, Clausen, T. | | Deposit date: | 2002-11-21 | | Release date: | 2002-12-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Determinants of Enzymatic Specificity in the Cys-Met-Metabolism PLP-Dependent Enzymes Family: Crystal Structure of Cystathionine gamma-lyase from Yeast and Intrafamiliar Structural Comparison
BIOL.CHEM., 384, 2003
|
|
1OXE
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
1J2P
 
 | | alpha-ring from the proteasome from archaeoglobus fulgidus | | Descriptor: | Proteasome alpha subunit | | Authors: | Groll, M, Brandstetter, H, Bartunik, H, Bourenkow, G, Huber, R. | | Deposit date: | 2003-01-08 | | Release date: | 2003-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigations on the Maturation and Regulation of Archaebacterial Proteasomes
J.MOL.BIOL., 327, 2003
|
|
1ITZ
 
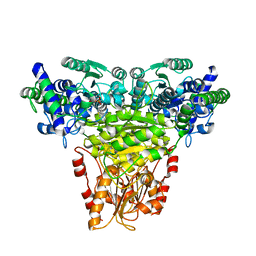 | | Maize Transketolase in complex with TPP | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, Transketolase | | Authors: | Gerhardt, S, Echt, S, Bader, G, Freigang, J, Busch, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-15 | | Release date: | 2003-02-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and properties of an engineered transketolase from maize
PLANT PHYSIOL., 132, 2003
|
|
1NBA
 
 | | CRYSTAL STRUCTURE ANALYSIS, REFINEMENT AND ENZYMATIC REACTION MECHANISM OF N-CARBAMOYLSARCOSINE AMIDOHYDROLASE FROM ARTHROBACTER SP. AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | N-CARBAMOYLSARCOSINE AMIDOHYDROLASE, SULFATE ION | | Authors: | Romao, M.J, Turk, D, Gomis-Ruth, F.-Z, Huber, R, Schumacher, G, Mollering, H, Russmann, L. | | Deposit date: | 1992-05-18 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure analysis, refinement and enzymatic reaction mechanism of N-carbamoylsarcosine amidohydrolase from Arthrobacter sp. at 2.0 A resolution.
J.Mol.Biol., 226, 1992
|
|
1OAH
 
 | | Cytochrome c Nitrite Reductase from Desulfovibrio desulfuricans ATCC 27774: The relevance of the two calcium sites in the structure of the catalytic subunit (NrfA). | | Descriptor: | CALCIUM ION, CHLORIDE ION, CYTOCHROME C NITRITE REDUCTASE, ... | | Authors: | Cunha, C.A, Macieira, S, Dias, J.M, Almeida, G, Goncalves, L.L, Costa, C, Lampreia, J, Huber, R, Moura, J.J.G, Moura, I, Romao, M.J. | | Deposit date: | 2003-01-14 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cytochrome C Nitrite Reductase from Desulfovibrio Desulfuricans Atcc 27774. The Relevance of the Two Calcium Sites in the Structure of the Catalytic Subunit (Nrfa)
J.Biol.Chem., 278, 2003
|
|
1J93
 
 | | Crystal Structure and Substrate Binding Modeling of the Uroporphyrinogen-III Decarboxylase from Nicotiana tabacum: Implications for the Catalytic Mechanism | | Descriptor: | SULFATE ION, UROPORPHYRINOGEN DECARBOXYLASE | | Authors: | Martins, B.M, Grimm, B, Mock, H.-P, Huber, R, Messerschmidt, A. | | Deposit date: | 2001-05-23 | | Release date: | 2001-10-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and substrate binding modeling of the uroporphyrinogen-III decarboxylase from Nicotiana tabacum. Implications for the catalytic mechanism
J.Biol.Chem., 276, 2001
|
|
1E4J
 
 | |
1M1G
 
 | | Crystal Structure of Aquifex aeolicus N-utilization substance G (NusG), Space Group P2(1) | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Steiner, T, Kaiser, J.T, Marinkovic, S, Huber, R, Wahl, M.C. | | Deposit date: | 2002-06-19 | | Release date: | 2003-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of transcription factor NusG in light of its nucleic
acid- and protein-binding activities
Embo J., 21, 2002
|
|
1N05
 
 | | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold | | Descriptor: | putative Riboflavin kinase | | Authors: | Bauer, S, Kemter, K, Bacher, A, Huber, R, Fischer, M, Steinbacher, S. | | Deposit date: | 2002-10-11 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold
J.Mol.Biol., 326, 2003
|
|
1ICP
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO COMPLEXED WITH PEG400 | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1CLV
 
 | | YELLOW MEAL WORM ALPHA-AMYLASE IN COMPLEX WITH THE AMARANTH ALPHA-AMYLASE INHIBITOR | | Descriptor: | CALCIUM ION, CHLORIDE ION, PROTEIN (ALPHA-AMYLASE INHIBITOR), ... | | Authors: | Pereira, P.J.B, Lozanov, V, Patthy, A, Huber, R, Bode, W, Pongor, S, Strobl, S. | | Deposit date: | 1999-05-04 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific inhibition of insect alpha-amylases: yellow meal worm alpha-amylase in complex with the amaranth alpha-amylase inhibitor at 2.0 A resolution.
Structure Fold.Des., 7, 1999
|
|
1N2T
 
 | | C-DES Mutant K223A with GLY Covalenty Linked to the PLP-cofactor | | Descriptor: | GLYCINE, L-cysteine/cystine lyase C-DES, POTASSIUM ION, ... | | Authors: | Kaiser, J.T, Bruno, S, Clausen, T, Huber, R, Schiaretti, F, Mozzarelli, A, Kessler, D. | | Deposit date: | 2002-10-24 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Snapshots of the Cystine Lyase "C-DES" during Catalysis: Studies in Solution and in the Crystalline State
J.Biol.Chem., 278, 2003
|
|
