1HLE
 
 | | CRYSTAL STRUCTURE OF CLEAVED EQUINE LEUCOCYTE ELASTASE INHIBITOR DETERMINED AT 1.95 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, HORSE LEUKOCYTE ELASTASE INHIBITOR | | Authors: | Baumann, U, Bode, W, Huber, R, Travis, J, Potempa, J. | | Deposit date: | 1992-04-13 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of cleaved equine leucocyte elastase inhibitor determined at 1.95 A resolution.
J.Mol.Biol., 226, 1992
|
|
1NPS
 
 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF PROTEIN S | | Descriptor: | CALCIUM ION, DEVELOPMENT-SPECIFIC PROTEIN S | | Authors: | Wenk, M, Baumgartner, R, Mayer, E.M, Huber, R, Holak, T.A, Jaenicke, R. | | Deposit date: | 1999-02-01 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The domains of protein S from Myxococcus xanthus: structure, stability and interactions.
J.Mol.Biol., 286, 1999
|
|
1EJN
 
 | | UROKINASE PLASMINOGEN ACTIVATOR B-CHAIN INHIBITOR COMPLEX | | Descriptor: | N-(1-ADAMANTYL)-N'-(4-GUANIDINOBENZYL)UREA, SULFATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Sperl, S, Jacob, U, Arroyo de Prada, N, Stuerzebecher, J, Wilhelm, O.G, Bode, W, Magdolen, V, Huber, R, Moroder, L. | | Deposit date: | 2000-04-22 | | Release date: | 2000-05-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | (4-aminomethyl)phenylguanidine derivatives as nonpeptidic highly selective inhibitors of human urokinase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1ORV
 
 | | Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Engel, M, Hoffmann, T, Wagner, L, Wermann, M, Heiser, U, Kiefersauer, R, Huber, R, Bode, W, Demuth, H.U, Brandstetter, H. | | Deposit date: | 2003-03-16 | | Release date: | 2003-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals its Functional Regulation and Enzymatic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1PX4
 
 | | E. COLI (LACZ) BETA-GALACTOSIDASE (G794A) WITH IPTG BOUND | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Juers, D.H, Hakda, S, Matthews, B.W, Huber, R.E. | | Deposit date: | 2003-07-02 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Altered Activity of Gly794 Variants of Escherichia coli Beta-Galactosidase
Biochemistry, 42, 2003
|
|
1PX3
 
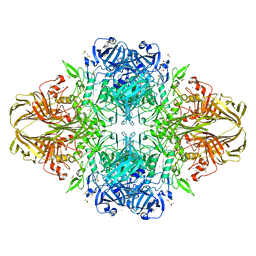 | | E. COLI (LACZ) BETA-GALACTOSIDASE (G794A) | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Juers, D.H, Hakda, S, Matthews, B.W, Huber, R.E. | | Deposit date: | 2003-07-02 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Altered Activity of Gly794 Variants of Escherichia coli Beta-Galactosidase
Biochemistry, 42, 2003
|
|
1YOJ
 
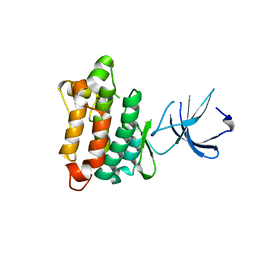 | | Crystal structure of Src kinase domain | | Descriptor: | proto-oncogene tyrosine-protein kinase SRC | | Authors: | Breitenlechner, C.B, Kairies, N.A, Honold, K, Scheiblich, S, Koll, H, Greiter, E, Koch, S, Schaefer, W, Huber, R, Engh, R.A. | | Deposit date: | 2005-01-27 | | Release date: | 2006-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of active SRC kinase domain complexes
J.Mol.Biol., 353, 2005
|
|
1PRC
 
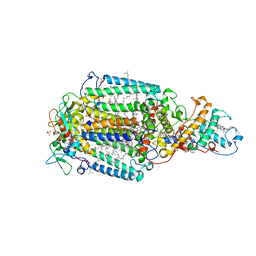 | | CRYSTALLOGRAPHIC REFINEMENT AT 2.3 ANGSTROMS RESOLUTION AND REFINED MODEL OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS | | Descriptor: | 15-trans-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Deisenhofer, J, Epp, O, Miki, K, Huber, R, Michel, H. | | Deposit date: | 1988-02-04 | | Release date: | 1989-01-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic refinement at 2.3 A resolution and refined model of the photosynthetic reaction centre from Rhodopseudomonas viridis.
J.Mol.Biol., 246, 1995
|
|
1YOL
 
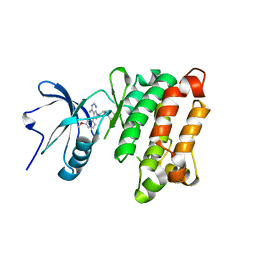 | | Crystal structure of Src kinase domain in complex with CGP77675 | | Descriptor: | 1-{4-[4-AMINO-5-(3-METHOXYPHENYL)-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL]BENZYL}PIPERIDIN-4-OL, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Breitenlechner, C.B, Kairies, N.A, Honold, K, Scheiblich, S, Koll, H, Greiter, E, Koch, S, Schaefer, W, Huber, R, Engh, R.A. | | Deposit date: | 2005-01-27 | | Release date: | 2006-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of active SRC kinase domain complexes
J.Mol.Biol., 353, 2005
|
|
1RFN
 
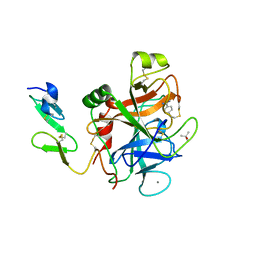 | | HUMAN COAGULATION FACTOR IXA IN COMPLEX WITH P-AMINO BENZAMIDINE | | Descriptor: | CALCIUM ION, P-AMINO BENZAMIDINE, PROTEIN (COAGULATION FACTOR IX), ... | | Authors: | Hopfner, K.-P, Lang, A, Karcher, A, Sichler, K, Kopetzki, E, Brandstetter, H, Huber, R, Bode, W, Engh, R.A. | | Deposit date: | 1999-04-19 | | Release date: | 1999-09-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Coagulation factor IXa: the relaxed conformation of Tyr99 blocks substrate binding.
Structure Fold.Des., 7, 1999
|
|
1NBA
 
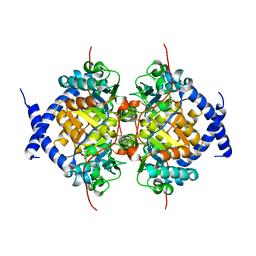 | | CRYSTAL STRUCTURE ANALYSIS, REFINEMENT AND ENZYMATIC REACTION MECHANISM OF N-CARBAMOYLSARCOSINE AMIDOHYDROLASE FROM ARTHROBACTER SP. AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | N-CARBAMOYLSARCOSINE AMIDOHYDROLASE, SULFATE ION | | Authors: | Romao, M.J, Turk, D, Gomis-Ruth, F.-Z, Huber, R, Schumacher, G, Mollering, H, Russmann, L. | | Deposit date: | 1992-05-18 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure analysis, refinement and enzymatic reaction mechanism of N-carbamoylsarcosine amidohydrolase from Arthrobacter sp. at 2.0 A resolution.
J.Mol.Biol., 226, 1992
|
|
1EHK
 
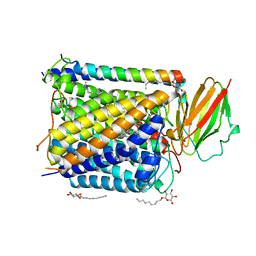 | | CRYSTAL STRUCTURE OF THE ABERRANT BA3-CYTOCHROME-C OXIDASE FROM THERMUS THERMOPHILUS | | Descriptor: | BA3-TYPE CYTOCHROME-C OXIDASE, COPPER (II) ION, DINUCLEAR COPPER ION, ... | | Authors: | Soulimane, T, Buse, G, Bourenkov, G.P, Bartunik, H.D, Huber, R, Than, M.E. | | Deposit date: | 2000-02-21 | | Release date: | 2001-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of the aberrant ba(3)-cytochrome c oxidase from thermus thermophilus.
EMBO J., 19, 2000
|
|
2B98
 
 | | Crystal Structure of an archaeal pentameric riboflavin synthase | | Descriptor: | Riboflavin synthase | | Authors: | Ramsperger, A, Augustin, M, Schott, A.K, Gerhardt, S, Krojer, T, Eisenreich, W, Illarionov, B, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of an Archaeal Pentameric Riboflavin Synthase in Complex with a Substrate Analog Inhibitor: stereochemical implications
J.Biol.Chem., 281, 2006
|
|
1ZLH
 
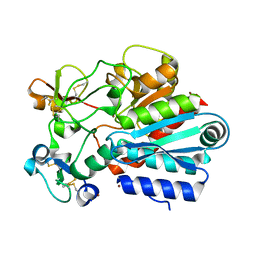 | | Crystal structure of the tick carboxypeptidase inhibitor in complex with bovine carboxypeptidase A | | Descriptor: | Carboxypeptidase A1, ZINC ION, carboxypeptidase inhibitor | | Authors: | Arolas, J.L, Popowicz, G.M, Lorenzo, J, Sommerhoff, C.P, Huber, R, Aviles, F.X, Holak, T.A. | | Deposit date: | 2005-05-06 | | Release date: | 2005-07-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Three-Dimensional Structures of Tick Carboxypeptidase Inhibitor in Complex with A/B Carboxypeptidases Reveal a Novel Double-headed Binding Mode
J.Mol.Biol., 350, 2005
|
|
1RVV
 
 | | SYNTHASE/RIBOFLAVIN SYNTHASE COMPLEX OF BACILLUS SUBTILIS | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, PHOSPHATE ION, RIBOFLAVIN SYNTHASE | | Authors: | Ritsert, K, Huber, R, Turk, D, Ladenstein, R, Schmidt-Baese, K, Bacher, A. | | Deposit date: | 1995-10-25 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Studies on the lumazine synthase/riboflavin synthase complex of Bacillus subtilis: crystal structure analysis of reconstituted, icosahedral beta-subunit capsids with bound substrate analogue inhibitor at 2.4 A resolution.
J.Mol.Biol., 253, 1995
|
|
1PFX
 
 | | PORCINE FACTOR IXA | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, FACTOR IXA | | Authors: | Brandstetter, H, Bauer, M, Huber, R, Lollar, P, Bode, W. | | Deposit date: | 1995-07-19 | | Release date: | 1996-08-17 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of clotting factor IXa: active site and module structure related to Xase activity and hemophilia B.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1PJP
 
 | | THE 2.2 A CRYSTAL STRUCTURE OF HUMAN CHYMASE IN COMPLEX WITH SUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYLKETONE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chymase, SUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYLKETONE INHIBITOR, ... | | Authors: | Pereira, P.J.B, Wang, Z.M, Rubin, H, Huber, R, Bode, W, Schechter, N.M, Strobl, S. | | Deposit date: | 1998-09-07 | | Release date: | 1999-03-02 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A crystal structure of human chymase in complex with succinyl-Ala-Ala-Pro-Phe-chloromethylketone: structural explanation for its dipeptidyl carboxypeptidase specificity.
J.Mol.Biol., 286, 1999
|
|
2ACH
 
 | | CRYSTAL STRUCTURE OF CLEAVED HUMAN ALPHA1-ANTICHYMOTRYPSIN AT 2.7 ANGSTROMS RESOLUTION AND ITS COMPARISON WITH OTHER SERPINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA 1-ANTICHYMOTRYPSIN, PHOSPHATE ION, ... | | Authors: | Baumann, U, Huber, R, Bode, W, Grosse, D, Lesjak, M, Laurell, C.B. | | Deposit date: | 1993-04-26 | | Release date: | 1993-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of cleaved human alpha 1-antichymotrypsin at 2.7 A resolution and its comparison with other serpins.
J.Mol.Biol., 218, 1991
|
|
1LI1
 
 | | The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link | | Descriptor: | ACETATE ION, Collagen alpha 1(IV), Collagen alpha 2(IV) | | Authors: | Than, M.E, Henrich, S, Huber, R, Ries, A, Mann, K, Kuhn, K, Timpl, R, Bourenkov, G.P, Bartunik, H.D, Bode, W. | | Deposit date: | 2002-04-17 | | Release date: | 2002-05-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1PO9
 
 | | Crytsal structure of isoaspartyl dipeptidase | | Descriptor: | Isoaspartyl dipeptidase, ZINC ION | | Authors: | Jozic, D, Kaiser, J.T, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2003-06-15 | | Release date: | 2004-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of isoaspartyl dipeptidase from E.coli: a dinuclear zinc peptidase evolved from amidohydrolases.
J.Mol.Biol., 332, 2003
|
|
1OBW
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE | | Descriptor: | INORGANIC PYROPHOSPHATASE, MAGNESIUM ION | | Authors: | Oganessyan, V.Yu, Harutyunyan, E.H, Avaeva, S.M, Oganessyan, N.N, Mather, T, Huber, R. | | Deposit date: | 1996-10-09 | | Release date: | 1997-09-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of holo inorganic pyrophosphatase from Escherichia coli at 1.9 A resolution. Mechanism of hydrolysis.
Biochemistry, 36, 1997
|
|
1NAS
 
 | | SEPIAPTERIN REDUCTASE COMPLEXED WITH N-ACETYL SEROTONIN | | Descriptor: | N-ACETYL SEROTONIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OXALOACETATE ION, ... | | Authors: | Auerbach, G, Herrmann, A, Bacher, A, Huber, R. | | Deposit date: | 1998-03-26 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 1.25 A crystal structure of sepiapterin reductase reveals its binding mode to pterins and brain neurotransmitters.
EMBO J., 16, 1997
|
|
2AZN
 
 | | X-RAY Structure of 2,5-diamino-6-ribosylamino-4(3h)-pyrimidinone 5-phosphate reductase | | Descriptor: | 2-(6-(2-CYCLOHEXYLETHOXY)-TETRAHYDRO-4,5-DIHYDROXY-2(HYDROXYMETHYL)-2H-PYRAN-3-YLOXY)-TETRAHYDRO-6(HYDROXYMETHYL)-2H-PY RAN-3,4,5-TRIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Chatwell, L, Bacher, A, Huber, R, Fischer, M, Krojer, T. | | Deposit date: | 2005-09-12 | | Release date: | 2006-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biosynthesis of riboflavin: structure and properties of 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5'-phosphate reductase of Methanocaldococcus jannaschii
J.Mol.Biol., 359, 2006
|
|
2B99
 
 | | Crystal Structure of an archaeal pentameric riboflavin synthase Complex with a Substrate analog inhibitor | | Descriptor: | 6,7-DIOXO-5H-8-RIBITYLAMINOLUMAZINE, Riboflavin synthase | | Authors: | Ramsperger, A, Augustin, M, Schott, A.K, Gerhardt, S, Krojer, T, Eisenreich, W, Illarionov, B, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of an Archaeal Pentameric Riboflavin Synthase in Complex with a Substrate Analog Inhibitor: stereochemical implications
J.Biol.Chem., 281, 2006
|
|
1POJ
 
 | | Isoaspartyl Dipeptidase with bound inhibitor | | Descriptor: | 2-{[[(1S)-1-AMINO-2-CARBOXYETHYL](DIHYDROXY)PHOSPHORANYL]METHYL}-4-METHYLPENTANOIC ACID, Isoaspartyl dipeptidase, ZINC ION | | Authors: | Jozic, D, Kaiser, J.T, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2003-06-15 | | Release date: | 2004-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray structure of isoaspartyl dipeptidase from E.coli: a dinuclear zinc peptidase evolved from amidohydrolases.
J.Mol.Biol., 332, 2003
|
|
