6IFE
 
 | | A Glycoside Hydrolase Family 43 beta-Xylosidase | | Descriptor: | Beta-xylosidase, GLYCEROL | | Authors: | Li, N, Liu, Y, Zhang, R, Zhou, J.P, Huang, Z.X. | | Deposit date: | 2018-09-20 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Biochemical and structural properties of a low-temperature-active glycoside hydrolase family 43 beta-xylosidase: Activity and instability at high neutral salt concentrations.
Food Chem, 301, 2019
|
|
5YHQ
 
 | | Cryo-EM Structure of CVA6 VLP | | Descriptor: | Capsid protein VP1, Capsid protein VP3, capsid protein VP0 | | Authors: | Chen, J, Zhang, C, Huang, Z, Cong, Y. | | Deposit date: | 2017-09-29 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A 3.0-Angstrom Resolution Cryo-Electron Microscopy Structure and Antigenic Sites of Coxsackievirus A6-Like Particles.
J. Virol., 92, 2018
|
|
8I9T
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State Dbp10-1 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8I9Z
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State Spb4 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8I9R
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State 5S RNP | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, 60S ribosomal protein L16-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8I9V
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State Dbp10-2 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8I9X
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - Ytm1-1 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8I9W
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - Dbp10-3 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8IA0
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State Puf6 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8I9Y
 
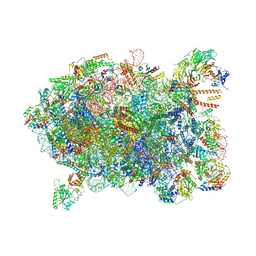 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - Ytm1-2 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
2F5H
 
 | | Solution structure of the alpha-domain of human Metallothionein-3 | | Descriptor: | CADMIUM ION, Metallothionein-3 | | Authors: | Wang, H, Zhang, Q, Cai, B, Li, H.Y, Sze, K.H, Huang, Z.X, Wu, H.M, Sun, H.Z. | | Deposit date: | 2005-11-25 | | Release date: | 2006-05-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of human metallothionein-3 (MT-3)
Febs Lett., 580, 2006
|
|
8IDZ
 
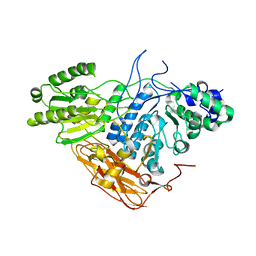 | |
8I12
 
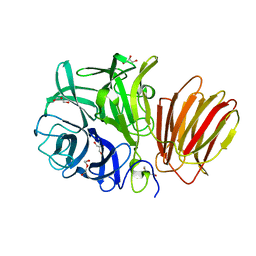 | | InuAMN8 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glycosyl hydrolase family 32 exo-inulinase | | Authors: | Zhou, J.P, Cen, X.L, He, L.M, Zhang, R, Huang, Z.X. | | Deposit date: | 2023-01-12 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Cold-active and NaCl-tolerant exo-inulinase InuAMN8.
To Be Published
|
|
7FJD
 
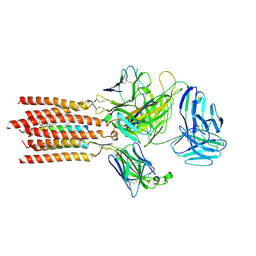 | | Cryo-EM structure of a membrane protein(WT) | | Descriptor: | CHOLESTEROL, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7FJE
 
 | | Cryo-EM structure of a membrane protein(LL) | | Descriptor: | CHOLESTEROL, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7FJF
 
 | | Cryo-EM structure of a membrane protein(CS) | | Descriptor: | CHOLEST-5-EN-3-YL HYDROGEN SULFATE, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7WSO
 
 | | Structure of a membrane protein G | | Descriptor: | B-cell antigen receptor complex-associated protein alpha chain, B-cell antigen receptor complex-associated protein beta chain, Immunoglobulin heavy constant gamma 1 | | Authors: | Ma, X, Zhu, Y, Chen, Y, Huang, Z. | | Deposit date: | 2022-01-30 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Cryo-EM structures of two human B cell receptor isotypes.
Science, 377, 2022
|
|
2LS4
 
 | |
2LS2
 
 | |
2LS3
 
 | |
5Y2F
 
 | | Human SIRT6 in complex with allosteric activator MDL-801 | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, 9-mer peptide QTARKSTGG, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, J, Huang, Z, Song, K. | | Deposit date: | 2017-07-25 | | Release date: | 2018-11-07 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification of a cellularly active SIRT6 allosteric activator.
Nat. Chem. Biol., 14, 2018
|
|
6IV6
 
 | | Cryo-EM structure of AcrVA5-acetylated MbCas12a in complex with crRNA | | Descriptor: | RNA (59-MER), nuclease | | Authors: | Dong, L, Li, N, Guan, X, Zhu, Y, Gao, N, Huang, Z. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An anti-CRISPR protein disables type V Cas12a by acetylation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
6KAF
 
 | | C2S2M2N2-type PSII-LHCII | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Chang, S.H, Shen, L.L, Huang, Z.H, Wang, W.D, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2019-06-22 | | Release date: | 2019-10-23 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structure of a C2S2M2N2-type PSII-LHCII supercomplex from the green algaChlamydomonas reinhardtii.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6AEG
 
 | | Crystal structure of xCas9 in complex with sgRNA and target DNA (GAT PAM) | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*AP*AP*GP*AP*TP*TP*AP*TP*TP*G)-3'), DNA nuclease, ... | | Authors: | Guo, M, Ren, K, Zhu, Y, Huang, Z. | | Deposit date: | 2018-08-04 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural insights into a high fidelity variant of SpCas9.
Cell Res., 29, 2019
|
|
6AEB
 
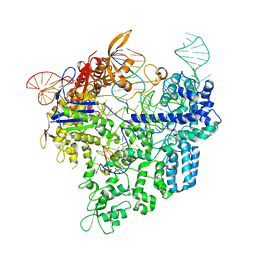 | | Crystal structure of xCas9 in complex with sgRNA and target DNA (AAG PAM) | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*AP*AP*AP*AP*GP*TP*AP*TP*TP*G)-3'), DNA Nuclease, ... | | Authors: | Guo, M, Ren, K, Zhu, Y, Huang, Z. | | Deposit date: | 2018-08-04 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structural insights into a high fidelity variant of SpCas9.
Cell Res., 29, 2019
|
|
