8H4J
 
 | | Crystal Structure of AzoR-FMN-Lyb24 complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN dependent NADH:quinone oxidoreductase, ~{N}-(3-methylsulfanylphenyl)-4~{H}-cyclopenta[b]quinolin-9-amine | | Authors: | Huang, W. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of AzoR-FMN-Lyb24 complex
To Be Published
|
|
8IND
 
 | | Crystal structure of UGT74AN3-UDP-RES | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, ... | | Authors: | Huang, W. | | Deposit date: | 2023-03-09 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8FZR
 
 | |
7UO0
 
 | | E.coli RNaseP Holoenzyme with Mg2+ | | Descriptor: | CALCIUM ION, Precursor tRNA substrate G(-1) G(-2), RNase P RNA, ... | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
7UO1
 
 | | E.coli RNaseP Holoenzyme with Mg2+ | | Descriptor: | CALCIUM ION, E.coli RNase P RNA, Precursor tRNA substrate U(-1) and A(-2), ... | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
7UO2
 
 | | E.coli RNaseP Holoenzyme with Mg2+ | | Descriptor: | MAGNESIUM ION, RNase P RNA, Ribonuclease P protein component | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
7UO5
 
 | | E.coli RNaseP Holoenzyme with Mg2+ | | Descriptor: | CALCIUM ION, RNase P RNA, Ribonuclease P protein component | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
7DUC
 
 | | Crystal Structure of cyto WalK | | Descriptor: | Sensor protein kinase WalK, ZINC ION | | Authors: | Huang, W. | | Deposit date: | 2021-01-08 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal Structure of cyto WalK
To Be Published
|
|
7DUD
 
 | | Crystal Structure of erWalK | | Descriptor: | Sensor protein kinase WalK | | Authors: | Huang, W. | | Deposit date: | 2021-01-08 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of erWalK
To Be Published
|
|
2JTC
 
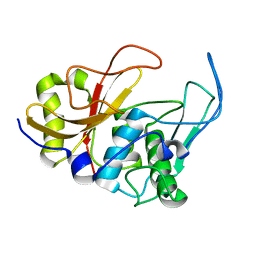 | | 3D structure and backbone dynamics of SPE B | | Descriptor: | Streptopain | | Authors: | Chuang, W, Wang, C, Houng, H, Chen, C, Wang, P. | | Deposit date: | 2007-07-26 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of streptopain: insight into diverse substrate specificity.
J.Biol.Chem., 284, 2009
|
|
2M75
 
 | |
2M7H
 
 | |
2M7F
 
 | |
2KIU
 
 | |
2B34
 
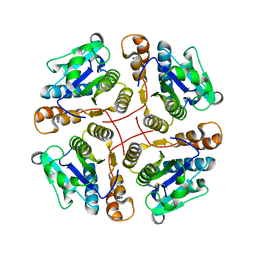 | | Structure of MAR1 Ribonuclease from Caenorhabditis elegans | | Descriptor: | MAR1 Ribonuclease | | Authors: | Schormann, N, Karpova, E, Li, S, Symersky, J, Zhang, Y, Lu, S, Zhou, Q, Lin, G, Cao, Z, Luo, M, Qiu, S, Luan, C.-H, Luo, D, Huang, W, Shang, Q, McKinstry, A, An, J, Tsao, J, Carson, M, Stinnett, M, Chen, Y, Johnson, D, Gary, R, Arabshahi, A, Bunzel, R, Bray, T, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-09-19 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structure of MAR1 Ribonuclease from Caenorhabditis elegans
To be Published
|
|
1AFR
 
 | |
8YRF
 
 | |
8INV
 
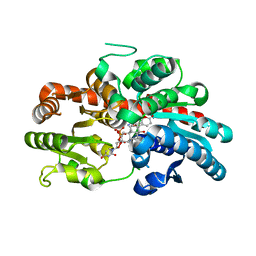 | | Crystal structure of UGT74AN3-UDP-BUF | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Long, F, Huang, W. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8K6Z
 
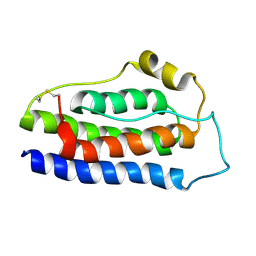 | | NMR structure of human leptin | | Descriptor: | Leptin | | Authors: | Fan, X, Qin, R, Yuan, W, Fan, J, Huang, W, Lin, Z. | | Deposit date: | 2023-07-26 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human leptin reveals a conformational plasticity important for receptor recognition.
Structure, 32, 2024
|
|
1EAP
 
 | | CRYSTAL STRUCTURE OF A CATALYTIC ANTIBODY WITH A SERINE PROTEASE ACTIVE SITE | | Descriptor: | IGG2B-KAPPA 17E8 FAB (HEAVY CHAIN), IGG2B-KAPPA 17E8 FAB (LIGHT CHAIN), PHENYL[1-(N-SUCCINYLAMINO)PENTYL]PHOSPHONATE | | Authors: | Zhou, G.W, Guo, J, Huang, W, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 1994-08-10 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a catalytic antibody with a serine protease active site.
Science, 265, 1994
|
|
8IR2
 
 | | Crystal structure of the SLF1 BRCT domain in complex with a Rad18 peptide containing pS442 and pS444 | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Xiang, S, Huang, W, Qiu, F. | | Deposit date: | 2023-03-17 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into Rad18 targeting by the SLF1 BRCT domains.
J.Biol.Chem., 299, 2023
|
|
1XDP
 
 | | Crystal Structure of the E.coli Polyphosphate Kinase in complex with AMPPNP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyphosphate kinase | | Authors: | Zhu, Y, Huang, W, Lee, S.S, Xu, W. | | Deposit date: | 2004-09-07 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a polyphosphate kinase and its implications for polyphosphate synthesis
Embo Rep., 6, 2005
|
|
6E53
 
 | | Structure of TERT in complex with a novel telomerase inhibitor | | Descriptor: | MAGNESIUM ION, RNA/DNA hairpin, Telomerase reverse transcriptase, ... | | Authors: | Hernandez-Sanchez, W, Huang, W, Plucinsky, B, Garcia-Vazquez, N, Berdis, A.J, Skordalakes, E, Taylor, D.J. | | Deposit date: | 2018-07-19 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A non-natural nucleotide uses a specific pocket to selectively inhibit telomerase activity.
Plos Biol., 17, 2019
|
|
1XDO
 
 | |
6UP7
 
 | | neurotensin receptor and arrestin2 complex | | Descriptor: | ARG-ARG-PRO-TYR-ILE-LEU, Beta-arrestin-1, Neurotensin receptor type 1, ... | | Authors: | Qu, Q.H, Huang, W, Masureel, M, Janetzko, J, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2019-10-16 | | Release date: | 2020-02-26 | | Last modified: | 2020-06-17 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the neurotensin receptor 1 in complex with beta-arrestin 1.
Nature, 579, 2020
|
|
