3WDW
 
 | | The apo-form structure of E113A from Paecilomyces thermophila | | Descriptor: | Beta-1,3-1,4-glucanase, SULFATE ION | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
3WDV
 
 | | The complex structure of PtLic16A with cellotetraose | | Descriptor: | Beta-1,3-1,4-glucanase, SULFATE ION, beta-D-glucopyranose, ... | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
3WDX
 
 | | The complex structure of E113A with glucotriose | | Descriptor: | Beta-1,3-1,4-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
7XGL
 
 | | Quinolinate Phosphoribosyl Transferase (QAPRTase) from Streptomyces pyridomyceticus NRRL B-2517 in Apo form | | Descriptor: | CHLORIDE ION, GLYCEROL, Quinolinate Phosphoribosyl Transferase, ... | | Authors: | Zhou, Z, Yang, X, Huang, T, Wang, X, Liang, R, Zheng, J, Dai, S, Lin, S, Deng, Z. | | Deposit date: | 2022-04-05 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Bifunctional NadC Homologue PyrZ Catalyzes Nicotinic Acid Formation in Pyridomycin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
7XGM
 
 | | Quinolinate Phosphoribosyl Transferase (QAPRTase) from Streptomyces pyridomyceticus NRRL B-2517 in complex with Quinolinic Acid (QA) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, QUINOLINIC ACID, ... | | Authors: | Zhou, Z, Yang, X, Huang, T, Wang, X, Liang, R, Zheng, J, Dai, S, Lin, S, Deng, Z. | | Deposit date: | 2022-04-05 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bifunctional NadC Homologue PyrZ Catalyzes Nicotinic Acid Formation in Pyridomycin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
7XGN
 
 | | Quinolinate Phosphoribosyl Transferase (QAPRTase) from Streptomyces pyridomyceticus NRRL B-2517 in complex with Nicotinic Acid (NA) | | Descriptor: | CHLORIDE ION, NICOTINIC ACID, Quinolinate Phosphoribosyl Transferase, ... | | Authors: | Zhou, Z, Yang, X, Huang, T, Wang, X, Liang, R, Zheng, J, Dai, S, Lin, S, Deng, Z. | | Deposit date: | 2022-04-05 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bifunctional NadC Homologue PyrZ Catalyzes Nicotinic Acid Formation in Pyridomycin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
3WP5
 
 | | The crystal structure of mutant CDBFV E109A from Neocallimastix patriciarum | | Descriptor: | CDBFV | | Authors: | Cheng, Y.S, Chen, C.C, Huang, C.H, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-01-09 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 11 xylanase from Neocallimastix patriciarum: insights into the molecular basis of a thermophilic enzyme.
J.Biol.Chem., 289, 2014
|
|
3WP4
 
 | | The crystal structure of native CDBFV from Neocallimastix patriciarum | | Descriptor: | CDBFV, SULFATE ION | | Authors: | Cheng, Y.S, Chen, C.C, Huang, C.H, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-01-09 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 11 xylanase from Neocallimastix patriciarum: insights into the molecular basis of a thermophilic enzyme.
J.Biol.Chem., 289, 2014
|
|
3WP6
 
 | | The complex structure of CDBFV E109A with xylotriose | | Descriptor: | CDBFV, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Cheng, Y.S, Chen, C.C, Huang, C.H, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-01-09 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 11 xylanase from Neocallimastix patriciarum: insights into the molecular basis of a thermophilic enzyme.
J.Biol.Chem., 289, 2014
|
|
5B7A
 
 | |
2WIB
 
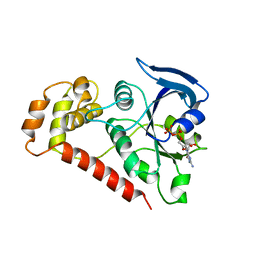 | | Crystal Structures of the N-terminal Intracellular Domain of FeoB from Klebsiella Pneumoniae in GDP binding state | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Hung, K.-W, Chang, Y.-W, Chen, J.-H, Chen, Y.-C, Sun, Y.-J, Hsiao, C.-D, Huang, T.-H. | | Deposit date: | 2009-05-09 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural Fold, Conservation and Fe(II) Binding of the Intracellular Domain of Prokaryote Feob.
J.Struct.Biol., 170, 2010
|
|
2WIA
 
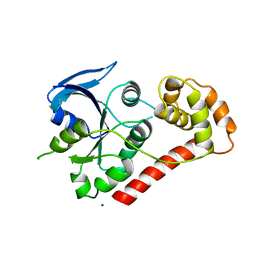 | | Crystal Structures of the N-terminal Intracellular Domain of FeoB from Klebsiella Pneumoniae in Apo Form | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B, MAGNESIUM ION | | Authors: | Hung, K.-W, Chang, Y.-W, Chen, J.-H, Chen, Y.-C, Sun, Y.-J, Hsiao, C.-D, Huang, T.-H. | | Deposit date: | 2009-05-09 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Fold, Conservation and Fe(II) Binding of the Intracellular Domain of Prokaryote Feob.
J.Struct.Biol., 170, 2010
|
|
2WIC
 
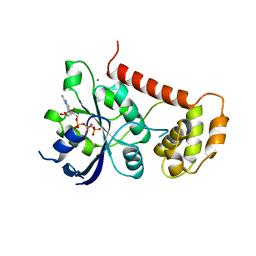 | | Crystal Structures of the N-terminal Intracellular Domain of FeoB from Klebsiella Pneumoniae in GMPPNP binding state | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Hung, K.-W, Chang, Y.-W, Chen, J.-H, Chen, Y.-C, Sun, Y.-J, Hsiao, C.-D, Huang, T.-H. | | Deposit date: | 2009-05-09 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Fold, Conservation and Fe(II) Binding of the Intracellular Domain of Prokaryote Feob.
J.Struct.Biol., 170, 2010
|
|
2KQS
 
 | |
2MVW
 
 | | Solution structure of the TRIM19 B-box1 (B1) of human promyelocytic leukemia (PML) | | Descriptor: | Protein PML, ZINC ION | | Authors: | Huang, S, Naik, M.T, Fan, P, Wang, Y, Chang, C, Huang, T. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The B-box 1 dimer of human promyelocytic leukemia protein.
J.Biomol.Nmr, 60, 2014
|
|
4AWX
 
 | | Moonlighting functions of FeoC in the regulation of ferrous iron transport in Feo | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B, FERROUS IRON TRANSPORT PROTEIN C, NICKEL (II) ION, ... | | Authors: | Hung, K.-W, Tsai, J.-Y, Juan, T.-H, Hsu, Y.-L, Hsiao, C.D, Huang, T.-H. | | Deposit date: | 2012-06-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Klebsiella Pneumoniae Nfeob/Feoc Complex and Roles of Feoc in Regulation of Fe2+ Transport by the Bacterial Feo System.
J.Bacteriol., 194, 2012
|
|
4NPN
 
 | | Crystal structure of human tetra-SUMO-2 | | Descriptor: | Small ubiquitin-related modifier 2 | | Authors: | Kung, C.C.-H, Naik, M.T, Chen, C.L, Ma, C, Huang, T.H. | | Deposit date: | 2013-11-22 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.633 Å) | | Cite: | Structural analysis of poly-SUMO chain recognition by the RNF4-SIMs domain.
Biochem.J., 462, 2014
|
|
3UZB
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
3UZO
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | Branched-chain-amino-acid aminotransferase, GLUTAMIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
4U92
 
 | | Crystal structure of a DNA/Ba2+ G-quadruplex containing a water-mediated C-tetrad | | Descriptor: | BARIUM ION, DNA (5'-D(*CP*CP*AP*KP*GP*CP*GP*TP*GP*G)-3'), MAGNESIUM ION | | Authors: | Paukstelis, P.J, Zhang, D, Huang, T, Lukeman, P. | | Deposit date: | 2014-08-05 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a DNA/Ba2+ G-quadruplex containing a water-mediated C-tetrad.
Nucleic Acids Res., 42, 2014
|
|
3UYY
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
1RI0
 
 | |
2JMH
 
 | | NMR solution structure of Blo t 5, a major mite allergen from Blomia tropicalis | | Descriptor: | Mite allergen Blo t 5 | | Authors: | Naik, M.T, Chang, C, Kuo, I, Chua, K, Huang, T. | | Deposit date: | 2006-11-12 | | Release date: | 2007-11-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Roles of Structure and Structural Dynamics in the Antibody Recognition of the Allergen Proteins: An NMR Study on Blomia tropicalis Major Allergen
Structure, 16, 2008
|
|
1PU3
 
 | |
3OSG
 
 | | The structure of protozoan parasite Trichomonas vaginalis Myb2 in complex with MRE-1-12 DNA | | Descriptor: | 5'-D(*AP*AP*AP*TP*AP*TP*CP*GP*TP*TP*AP*T)-3', 5'-D(*AP*TP*AP*AP*CP*GP*AP*TP*AP*TP*TP*T)-3', MYB21 | | Authors: | Jiang, I, Tsai, C.K, Chen, S.C, Wang, S.H, Amiraslanov, I, Chang, C.F, Wu, W.J, Tai, J.H, Liaw, Y.C, Huang, T.H. | | Deposit date: | 2010-09-09 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Molecular basis of the recognition of the ap65-1 gene transcription promoter elements by a Myb protein from the protozoan parasite Trichomonas vaginalis.
Nucleic Acids Res., 39, 2011
|
|
