1KOP
 
 | | NEISSERIA GONORRHOEAE CARBONIC ANHYDRASE | | Descriptor: | AZIDE ION, BETA-MERCAPTOETHANOL, CARBONIC ANHYDRASE, ... | | Authors: | Huang, S, Xue, Y, Chirica, L, Lindskog, S, Jonsson, B.-H. | | Deposit date: | 1998-03-22 | | Release date: | 1998-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of carbonic anhydrase from Neisseria gonorrhoeae and its complex with the inhibitor acetazolamide.
J.Mol.Biol., 283, 1998
|
|
6LKY
 
 | |
1M3W
 
 | | Crystal Structure of a Molecular Maquette Scaffold | | Descriptor: | H10H24, MERCURY (II) ION | | Authors: | Huang, S.S, Gibney, B.R, Stayrook, S.E, Dutton, P.L, Lewis, M. | | Deposit date: | 2002-07-01 | | Release date: | 2003-02-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Structure of a Maquette Scaffold
J.Mol.Biol., 326, 2003
|
|
4ESX
 
 | | Crystal structure of C. albicans Thi5 complexed with PLP | | Descriptor: | Pyrimidine biosynthesis enzyme THI13 | | Authors: | Huang, S, Fenwick, M.K, Zhang, Y, Lai, R, Hazra, A, Rajashankar, K, Philmus, B, Kinsland, C, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-04-23 | | Release date: | 2012-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Thiamin pyrimidine biosynthesis in Candida albicans : a remarkable reaction between histidine and pyridoxal phosphate.
J.Am.Chem.Soc., 134, 2012
|
|
7XTB
 
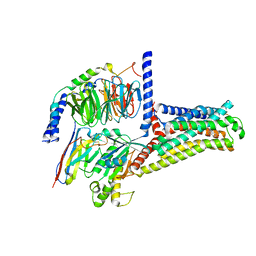 | | Serotonin 6 (5-HT6) receptor-Gs-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7XTC
 
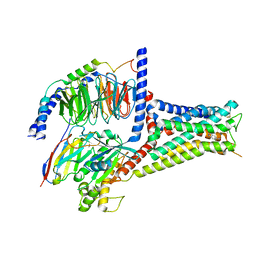 | | Serotonin 7 (5-HT7) receptor-Gs-Nb35 complex | | Descriptor: | 3-(2-azanylethyl)-1H-indole-5-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7XTA
 
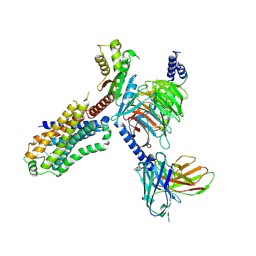 | | Serotonin 4 (5-HT4) receptor-Gi-scFv16 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7XT9
 
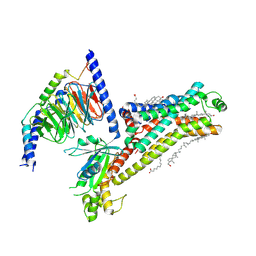 | | Serotonin 4 (5-HT4) receptor-Gs complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7XT8
 
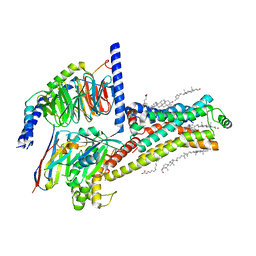 | | Serotonin 4 (5-HT4) receptor-Gs-Nb35 complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
4GIL
 
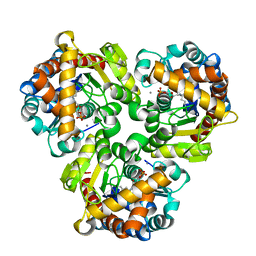 | | Crystal Structure of Pseudouridine Monophosphate Glycosidase/Linear Pseudouridine 5'-Phosphate Adduct | | Descriptor: | MANGANESE (II) ION, Pseudouridine-5'-phosphate glycosidase, pseudouridine 5'-phosphate, ... | | Authors: | Huang, S, Mahanta, N, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.539 Å) | | Cite: | Pseudouridine monophosphate glycosidase: a new glycosidase mechanism.
Biochemistry, 51, 2012
|
|
4GIM
 
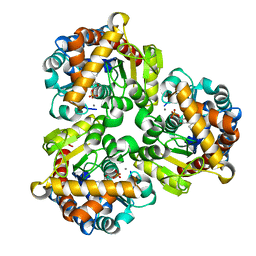 | | Crystal Structure of Pseudouridine Monophosphate Glycosidase Complexed with Pseudouridine 5'-phosphate | | Descriptor: | MANGANESE (II) ION, PSEUDOURIDINE-5'-MONOPHOSPHATE, Pseudouridine-5'-phosphate glycosidase | | Authors: | Huang, S, Mahanta, N, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Pseudouridine monophosphate glycosidase: a new glycosidase mechanism.
Biochemistry, 51, 2012
|
|
4GIJ
 
 | | Crystal Structure of Pseudouridine Monophosphate Glycosidase Complexed with Sulfate | | Descriptor: | MANGANESE (II) ION, Pseudouridine-5'-phosphate glycosidase, SULFATE ION | | Authors: | Huang, S, Mahanta, N, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Pseudouridine monophosphate glycosidase: a new glycosidase mechanism.
Biochemistry, 51, 2012
|
|
4GIK
 
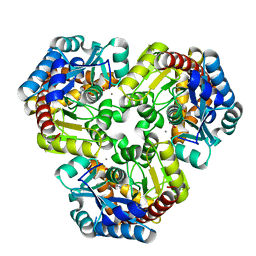 | | Crystal Structure of Pseudouridine Monophosphate Glycosidase/Linear R5P Adduct | | Descriptor: | MANGANESE (II) ION, Pseudouridine-5'-phosphate glycosidase, RIBOSE-5-PHOSPHATE | | Authors: | Huang, S, Mahanta, N, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Pseudouridine monophosphate glycosidase: a new glycosidase mechanism.
Biochemistry, 51, 2012
|
|
8JPT
 
 | |
8JPU
 
 | |
3UCJ
 
 | | Coccomyxa beta-carbonic anhydrase in complex with acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CHLORIDE ION, Carbonic anhydrase, ... | | Authors: | Huang, S, Hainzl, T, Sauer-Eriksson, A.E. | | Deposit date: | 2011-10-27 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies of [beta]-carbonic anhydrase from the green alga Coccomyxa: inhibitor complexes with anions and acetazolamide.
Plos One, 6, 2011
|
|
3UCN
 
 | | Coccomyxa beta-carbonic anhydrase in complex with azide | | Descriptor: | AZIDE ION, CHLORIDE ION, Carbonic anhydrase, ... | | Authors: | Huang, S, Hainzl, T, Sauer-Eriksson, A.E. | | Deposit date: | 2011-10-27 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural studies of [beta]-carbonic anhydrase from the green alga Coccomyxa: inhibitor complexes with anions and acetazolamide.
Plos One, 6, 2011
|
|
3UCO
 
 | |
3UCM
 
 | |
3UCK
 
 | | Coccomyxa beta-carbonic anhydrase in complex with phosphate | | Descriptor: | CHLORIDE ION, Carbonic anhydrase, PHOSPHATE ION, ... | | Authors: | Huang, S, Hainzl, T, Sauer-Eriksson, A.E. | | Deposit date: | 2011-10-27 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of [beta]-carbonic anhydrase from the green alga Coccomyxa: inhibitor complexes with anions and acetazolamide.
Plos One, 6, 2011
|
|
1KOQ
 
 | | NEISSERIA GONORRHOEAE CARBONIC ANHYDRASE | | Descriptor: | CARBONIC ANHYDRASE, ZINC ION | | Authors: | Huang, S, Xue, Y, Chirica, L, Lindskog, S, Jonsson, B.-H. | | Deposit date: | 1998-03-22 | | Release date: | 1998-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of carbonic anhydrase from Neisseria gonorrhoeae and its complex with the inhibitor acetazolamide.
J.Mol.Biol., 283, 1998
|
|
8IA9
 
 | |
8IAA
 
 | |
1LG5
 
 | | Crystal Structure Analysis of the HCA II Mutant T199P in complex with beta-mercaptoethanol | | Descriptor: | BETA-MERCAPTOETHANOL, Carbonic anhydrase II, ZINC ION | | Authors: | Huang, S, Sjoblom, B, Sauer-Eriksson, A.E, Jonsson, B.-H. | | Deposit date: | 2002-04-15 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Organization of an efficient carbonic anhydrase: implications for the mechanism based on structure-function studies of a T199P/C206S mutant.
Biochemistry, 41, 2002
|
|
1LGD
 
 | | Crystal Structure Analysis of HCA II Mutant T199P in Complex with Bicarbonate | | Descriptor: | BICARBONATE ION, Carbonic anhydrase II, ZINC ION | | Authors: | Huang, S, Sjoblom, B, Sauer-Eriksson, A.E, Jonsson, B.-H. | | Deposit date: | 2002-04-15 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Organization of an efficient carbonic anhydrase: implications for the mechanism based on structure-function studies of a T199P/C206S mutant.
Biochemistry, 41, 2002
|
|
