5LR3
 
 | |
5LR5
 
 | |
5NDI
 
 | |
5NEP
 
 | | The structure of the G. violaceus guanidine II riboswitch P1 stem-loop with methylguanidine | | Descriptor: | 1-METHYLGUANIDINE, RNA (5'-R(*GP*GP*UP*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*AP*(CBV)P*C)-3'), SODIUM ION, ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-03-11 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NY8
 
 | | The structure of the thermobifida fusca guanidine III riboswitch with aminoguanidine | | Descriptor: | AMINOGUANIDINE, MAGNESIUM ION, RNA (41-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NZ6
 
 | |
5LQO
 
 | |
5O62
 
 | |
5O69
 
 | | The structure of the thermobifida fusca guanidine III riboswitch with agmatine. | | Descriptor: | AGMATINE, MAGNESIUM ION, RNA (37-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-06-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
1LFD
 
 | | CRYSTAL STRUCTURE OF THE ACTIVE RAS PROTEIN COMPLEXED WITH THE RAS-INTERACTING DOMAIN OF RALGDS | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RALGDS, ... | | Authors: | Huang, L, Hofer, F, Martin, G.S, Kim, S.-H. | | Deposit date: | 1998-04-29 | | Release date: | 1999-05-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the interaction of Ras with RalGDS.
Nat.Struct.Biol., 5, 1998
|
|
6Q8V
 
 | |
1YQ4
 
 | | Avian respiratory complex ii with 3-nitropropionate and ubiquinone | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, 3-NITROPROPANOIC ACID, Coenzyme Q10, ... | | Authors: | Huang, L, Sun, G, Cobessi, D, Wang, A, Shen, J.T, Tung, E.Y, Anderson, V.E, Berry, E.A. | | Deposit date: | 2005-02-01 | | Release date: | 2005-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | 3-Nitropropionic Acid Is a Suicide Inhibitor of Mitochondrial Respiration That, upon Oxidation by Complex II, Forms a Covalent Adduct with a Catalytic Base Arginine in the Active Site of the Enzyme
J.Biol.Chem., 281, 2006
|
|
1YQ3
 
 | | Avian respiratory complex ii with oxaloacetate and ubiquinone | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Coenzyme Q10, (2Z,6E,10Z,14E,18E,22E,26Z)-isomer, ... | | Authors: | Huang, L, Cobessi, D, Tung, E.Y, Berry, E.A. | | Deposit date: | 2005-02-01 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3-Nitropropionic Acid Is a Suicide Inhibitor of Mitochondrial Respiration That, upon Oxidation by Complex II, Forms a Covalent Adduct with a Catalytic Base Arginine in the Active Site of the Enzyme
J.Biol.Chem., 281, 2006
|
|
1C4Z
 
 | | STRUCTURE OF AN E6AP-UBCH7 COMPLEX: INSIGHTS INTO THE UBIQUITINATION PATHWAY | | Descriptor: | UBIQUITIN CONJUGATING ENZYME E2, UBIQUITIN-PROTEIN LIGASE E3A | | Authors: | Huang, L, Kinnucan, E, Wang, G, Beaudenon, S, Howley, P.M, Huibregtse, J.M, Pavletich, N.P. | | Deposit date: | 1999-10-14 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade.
Science, 286, 1999
|
|
1LXD
 
 | | CRYSTAL STRUCTURE OF THE RAS INTERACTING DOMAIN OF RALGDS, A GUANINE NUCLEOTIDE DISSOCIATION STIMULATOR OF RAL PROTEIN | | Descriptor: | RALGDSB | | Authors: | Huang, L, Weng, X.W, Hofer, F, Martin, G.S, Kim, S.H. | | Deposit date: | 1997-03-05 | | Release date: | 1998-03-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure of the Ras-interacting domain of RalGDS.
Nat.Struct.Biol., 4, 1997
|
|
6YL5
 
 | | Crystal structure of the SAM-SAH riboswitch with SAH | | Descriptor: | Chains: A,B,C,D,E,F,G,H,I,J,K,L, MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 2020
|
|
3CWB
 
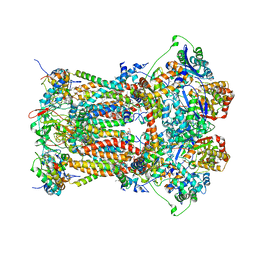 | | Chicken Cytochrome BC1 Complex inhibited by an iodinated analogue of the polyketide Crocacin-D | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, AZIDE ION, CARDIOLIPIN, ... | | Authors: | Huang, L, Cromartie, T, Viner, R, Crowley, P.J, Berry, E.A. | | Deposit date: | 2008-04-21 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | The role of molecular modeling in the design of analogues of the fungicidal natural products crocacins A and D.
Bioorg.Med.Chem., 16, 2008
|
|
6YMI
 
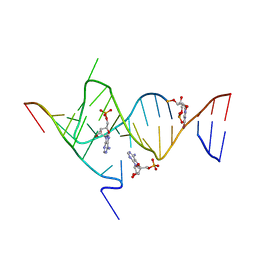 | | Crystal structure of the SAM-SAH riboswitch with AMP. | | Descriptor: | 5-BROMOCYTIDINE 5'-(DIHYDROGEN PHOSPHATE), ADENOSINE MONOPHOSPHATE, Chains: A,C,F,I,M,O, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
6YMJ
 
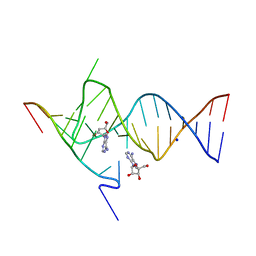 | | Crystal structure of the SAM-SAH riboswitch with adenosine. | | Descriptor: | 5-BROMOCYTIDINE 5'-(DIHYDROGEN PHOSPHATE), ADENOSINE, Chains: A,C,F,I,M,O, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
6YLB
 
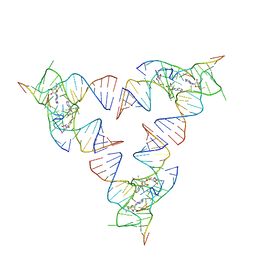 | | Crystal structure of the SAM-SAH riboswitch with SAM | | Descriptor: | Chains: A,C,F,I,M,O, Chains: B,D,G,J,N,P, S-ADENOSYLMETHIONINE | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 2020
|
|
6YMK
 
 | | Crystal structure of the SAM-SAH riboswitch with AMP | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Chains: A,C,F,I,M,O, Chains: B,D,G,J,N,P, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
5LQT
 
 | |
2H89
 
 | | Avian Respiratory Complex II with Malonate Bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Huang, L.S, Shen, J.T, Wang, A.C, Berry, E.A. | | Deposit date: | 2006-06-06 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the
Biochim.Biophys.Acta, 1757
|
|
6YML
 
 | |
6YMM
 
 | |
