3WP6
 
 | | The complex structure of CDBFV E109A with xylotriose | | Descriptor: | CDBFV, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Cheng, Y.S, Chen, C.C, Huang, C.H, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-01-09 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 11 xylanase from Neocallimastix patriciarum: insights into the molecular basis of a thermophilic enzyme.
J.Biol.Chem., 289, 2014
|
|
3WP4
 
 | | The crystal structure of native CDBFV from Neocallimastix patriciarum | | Descriptor: | CDBFV, SULFATE ION | | Authors: | Cheng, Y.S, Chen, C.C, Huang, C.H, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-01-09 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 11 xylanase from Neocallimastix patriciarum: insights into the molecular basis of a thermophilic enzyme.
J.Biol.Chem., 289, 2014
|
|
3WP5
 
 | | The crystal structure of mutant CDBFV E109A from Neocallimastix patriciarum | | Descriptor: | CDBFV | | Authors: | Cheng, Y.S, Chen, C.C, Huang, C.H, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-01-09 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 11 xylanase from Neocallimastix patriciarum: insights into the molecular basis of a thermophilic enzyme.
J.Biol.Chem., 289, 2014
|
|
8K32
 
 | |
4QVG
 
 | | Crystal structure of S-adenosylmethionine-dependent methyltransferase SibL in its apo form | | Descriptor: | SibL | | Authors: | Liu, J.S, Chen, S.C, Huang, C.H, Yang, C.S, Chen, Y. | | Deposit date: | 2014-07-15 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and mechanism of an antibiotics-synthesizing 3-hydroxykynurenine C-methyltransferase
Sci Rep, 5, 2015
|
|
4KT8
 
 | | The complex structure of Rv3378c-Y51FY90F with substrate, TPP | | Descriptor: | (2E)-3-methyl-5-[(1R,2S,8aS)-1,2,5,5-tetramethyl-1,2,3,5,6,7,8,8a-octahydronaphthalen-1-yl]pent-2-en-1-yl trihydrogen diphosphate, Diterpene synthase, PHOSPHATE ION | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
3WZL
 
 | | ZEN lactonase | | Descriptor: | Zearalenone hydrolase | | Authors: | Ko, T.P, Huang, C.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and substrate-binding mode of the mycoestrogen-detoxifying lactonase ZHD from Clonostachys rosea
RSC ADV, 4, 2014
|
|
3WZM
 
 | | ZEN lactonase mutant complex | | Descriptor: | (3S,11E)-14,16-dihydroxy-3-methyl-3,4,5,6,9,10-hexahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Zearalenone hydrolase | | Authors: | Ko, T.P, Huang, C.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure and substrate-binding mode of the mycoestrogen-detoxifying lactonase ZHD from Clonostachys rosea
RSC ADV, 4, 2014
|
|
3VNI
 
 | | Crystal structures of D-Psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars | | Descriptor: | MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-16 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3W01
 
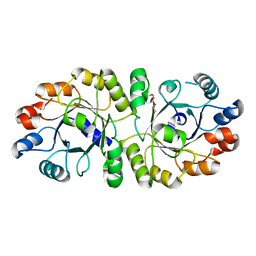 | | Crystal structure of PcrB complexed with PEG from Staphylococcus aureus subsp. aureus Mu3 | | Descriptor: | Heptaprenylglyceryl phosphate synthase, TRIETHYLENE GLYCOL | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VNJ
 
 | | Crystal structures of D-Psicose 3-epimerase with D-psicose from Clostridium cellulolyticum H10 | | Descriptor: | D-psicose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-16 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VZZ
 
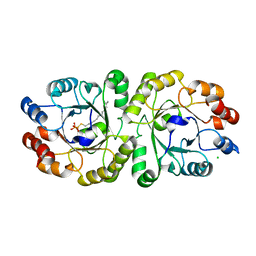 | | Crystal structure of PcrB complexed with FsPP from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VZX
 
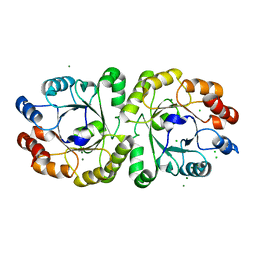 | | Crystal structure of PcrB from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VMV
 
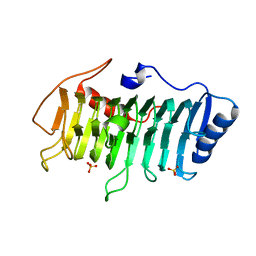 | | Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 | | Descriptor: | Pectate lyase, SULFATE ION | | Authors: | Zheng, Y, Huang, C.H, Liu, W, Ko, T.P, Xue, Y, Zhou, C, Zhang, G, Guo, R.T, Ma, Y. | | Deposit date: | 2011-12-16 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure and substrate-binding mode of a novel pectate lyase from alkaliphilic Bacillus sp. N16-5.
Biochem.Biophys.Res.Commun., 420, 2012
|
|
3VNM
 
 | | Crystal structures of D-Psicose 3-epimerase with D-sorbose from Clostridium cellulolyticum H10 | | Descriptor: | D-sorbose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-17 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VNK
 
 | | Crystal structures of D-Psicose 3-epimerase with D-fructose from Clostridium cellulolyticum H10 | | Descriptor: | D-fructose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-16 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VZY
 
 | | Crystal structure of PcrB complexed with G1P from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VMW
 
 | | Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate | | Descriptor: | Pectate lyase, SULFATE ION, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Zheng, Y, Huang, C.H, Liu, W, Ko, T.P, Xue, Y, Zhou, C, Zhang, G, Guo, R.T, Ma, Y. | | Deposit date: | 2011-12-17 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and substrate-binding mode of a novel pectate lyase from alkaliphilic Bacillus sp. N16-5.
Biochem.Biophys.Res.Commun., 420, 2012
|
|
3VNL
 
 | | Crystal structures of D-Psicose 3-epimerase with D-tagatose from Clostridium cellulolyticum H10 | | Descriptor: | D-tagatose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-16 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3W00
 
 | | Crystal structure of PcrB complexed with G1P and FsPP from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | Heptaprenylglyceryl phosphate synthase, PHOSPHATE ION, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3WL8
 
 | | Crystal Structure of pOPH S172A with octanoic acid | | Descriptor: | CITRIC ACID, OCTANOIC ACID (CAPRYLIC ACID), Oxidized polyvinyl alcohol hydrolase | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chan, H.C, Ren, F.F, Jia, D.X, Wang, A.H.-J, Guo, R.T, Chen, J, Du, G.C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into enzymatic degradation of oxidized polyvinyl alcohol
Chembiochem, 15, 2014
|
|
3WUB
 
 | | The wild type crystal structure of b-1,4-Xylanase (XynAS9) from Streptomyces sp. 9 | | Descriptor: | Endo-1,4-beta-xylanase A, ZINC ION | | Authors: | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
3WCH
 
 | | The complex structure of HsSQS wtih ligand BPH1237 | | Descriptor: | Squalene synthase, hydrogen [(1R)-2-(3-decyl-1H-imidazol-3-ium-1-yl)-1-hydroxy-1-phosphonoethyl]phosphonate | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
3WGZ
 
 | | Crystal structure of meso-dapdh Q154L/T173I/R199M/P248S/H249N/N276S mutant with D-leucine of from Clostridium tetani E88 | | Descriptor: | D-LEUCINE, Meso-diaminopimelate dehydrogenase | | Authors: | Liu, W.D, Li, Z, Huang, C.H, Guo, R.T, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2013-08-20 | | Release date: | 2014-08-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of meso-dapdh Q154L/T173I/R199M/P248S/H249N/N276S mutant with D-leucine of from Clostridium tetani E88
to be published
|
|
3WC9
 
 | | The complex structure of HsSQS wtih ligand, FSPP | | Descriptor: | S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, Squalene synthase | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-26 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
