8UX1
 
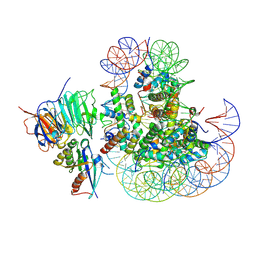 | |
8XDD
 
 | | Cryo-EM structure of human urea transporter A2. | | Descriptor: | 1-(3-methoxyphenyl)methanamine, 8-hydroxyquinoline-2-carboxylic acid, Urea transporter 2 | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
8XDG
 
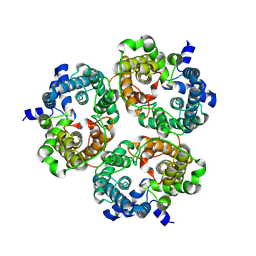 | |
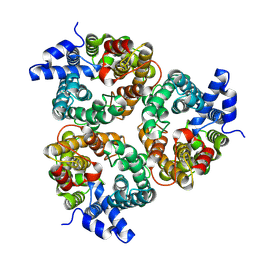
8XDE
 
 | |
8XDA
 
 | | Cryo-EM structure of urea bound human urea transporter A2. | | Descriptor: | UREA, Urea transporter 2 | | Authors: | Huang, S, Liu, L, Sun, J, Zhizheng, H. | | Deposit date: | 2023-12-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
8XDH
 
 | |
8XD9
 
 | |
8XD7
 
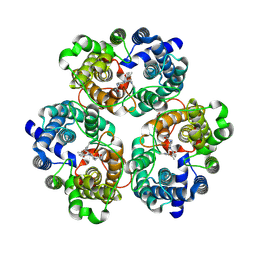 | |
8XDF
 
 | |
8XDC
 
 | | Cryo-EM structure of human urea transporter A2. | | Descriptor: | N-[3-[1,1-bis(oxidanylidene)-1,2-thiazolidin-2-yl]-4-chloranyl-phenyl]-2-methoxy-5-methyl-benzenesulfonamide, Urea transporter 2 | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
8XDI
 
 | | Cryo-EM structure of zebrafish urea transporter. | | Descriptor: | N-(4-acetamidophenyl)-5-ethanoyl-furan-2-carboxamide, Urea transporter | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-11 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
1LGD
 
 | | Crystal Structure Analysis of HCA II Mutant T199P in Complex with Bicarbonate | | Descriptor: | BICARBONATE ION, Carbonic anhydrase II, ZINC ION | | Authors: | Huang, S, Sjoblom, B, Sauer-Eriksson, A.E, Jonsson, B.-H. | | Deposit date: | 2002-04-15 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Organization of an efficient carbonic anhydrase: implications for the mechanism based on structure-function studies of a T199P/C206S mutant.
Biochemistry, 41, 2002
|
|
9IMP
 
 | | The complex of PDZ3 and PBM | | Descriptor: | INSC spindle orientation adaptor protein, Partitioning defective 3 homolog | | Authors: | Huang, S.J. | | Deposit date: | 2024-07-04 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | mInsc coordinates Par3 and NuMA condensates for assembly of the spindle orientation machinery in asymmetric cell division.
Int.J.Biol.Macromol., 279, 2024
|
|
8XDB
 
 | | Cryo-EM structure of human urea transporter A2. | | Descriptor: | (5E)-2-azanylidene-5-[(2,3-dimethoxyphenyl)methylidene]-1,3-thiazolidin-4-one, Urea transporter 2 | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
8YN1
 
 | | Cryo-EM structure of NRG1A(LRR) in complex with EDS1-SAG101-(ADPr-ATP) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5-DIPHOSPHORIBOSE, Probable disease resistance protein At5g66900, ... | | Authors: | Huang, S, Xiao, Y, Chai, J. | | Deposit date: | 2024-03-10 | | Release date: | 2024-12-11 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Balanced plant helper NLR activation by a modified host protein complex.
Nature, 639, 2025
|
|
8YN0
 
 | | Crystal structure of NRG1C in complex with EDS1-SAG101-(ADPr-ATP) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5-DIPHOSPHORIBOSE, Probable disease resistance protein At5g66890, ... | | Authors: | Huang, S, Xiao, Y, Chai, J. | | Deposit date: | 2024-03-10 | | Release date: | 2024-12-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Balanced plant helper NLR activation by a modified host protein complex.
Nature, 639, 2025
|
|
1LG5
 
 | | Crystal Structure Analysis of the HCA II Mutant T199P in complex with beta-mercaptoethanol | | Descriptor: | BETA-MERCAPTOETHANOL, Carbonic anhydrase II, ZINC ION | | Authors: | Huang, S, Sjoblom, B, Sauer-Eriksson, A.E, Jonsson, B.-H. | | Deposit date: | 2002-04-15 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Organization of an efficient carbonic anhydrase: implications for the mechanism based on structure-function studies of a T199P/C206S mutant.
Biochemistry, 41, 2002
|
|
1LG6
 
 | | Crystal Structure Analysis of HCA II Mutant T199P in Complex with Thiocyanate | | Descriptor: | Carbonic anhydrase II, THIOCYANATE ION, ZINC ION | | Authors: | Huang, S, Sjoblom, B, Sauer-Eriksson, A.E, Jonsson, B.-H. | | Deposit date: | 2002-04-15 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Organization of an efficient carbonic anhydrase: implications for the mechanism based on structure-function studies of a T199P/C206S mutant.
Biochemistry, 41, 2002
|
|
1KOQ
 
 | | NEISSERIA GONORRHOEAE CARBONIC ANHYDRASE | | Descriptor: | CARBONIC ANHYDRASE, ZINC ION | | Authors: | Huang, S, Xue, Y, Chirica, L, Lindskog, S, Jonsson, B.-H. | | Deposit date: | 1998-03-22 | | Release date: | 1998-12-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of carbonic anhydrase from Neisseria gonorrhoeae and its complex with the inhibitor acetazolamide.
J.Mol.Biol., 283, 1998
|
|
8JPT
 
 | |
8JOH
 
 | |
1KOP
 
 | | NEISSERIA GONORRHOEAE CARBONIC ANHYDRASE | | Descriptor: | AZIDE ION, BETA-MERCAPTOETHANOL, CARBONIC ANHYDRASE, ... | | Authors: | Huang, S, Xue, Y, Chirica, L, Lindskog, S, Jonsson, B.-H. | | Deposit date: | 1998-03-22 | | Release date: | 1998-12-09 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of carbonic anhydrase from Neisseria gonorrhoeae and its complex with the inhibitor acetazolamide.
J.Mol.Biol., 283, 1998
|
|
5GPE
 
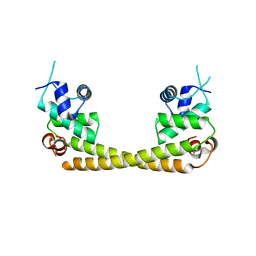 | | Crystal structure of the transcription regulator PbrR691 from Ralstonia metallidurans CH34 in complex with Lead(II) | | Descriptor: | LEAD (II) ION, Transcriptional regulator, MerR-family | | Authors: | Huang, S.Q, Chen, W.Z, Wang, D, Hu, Q.Y, Liu, X.C, Gan, J.H, Chen, H. | | Deposit date: | 2016-08-01 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis for the Selective Pb(II) Recognition of Metalloregulatory Protein PbrR691
Inorg Chem, 55, 2016
|
|
2MVW
 
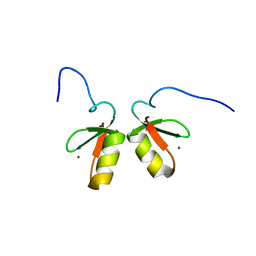 | | Solution structure of the TRIM19 B-box1 (B1) of human promyelocytic leukemia (PML) | | Descriptor: | Protein PML, ZINC ION | | Authors: | Huang, S, Naik, M.T, Fan, P, Wang, Y, Chang, C, Huang, T. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The B-box 1 dimer of human promyelocytic leukemia protein.
J.Biomol.Nmr, 60, 2014
|
|
8JPU
 
 | |
