4W98
 
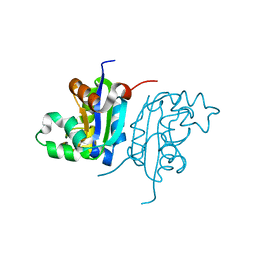 | | Acinetobacter baumannii SDF NDK | | 分子名称: | Nucleoside diphosphate kinase | | 著者 | Hu, Y, Feng, F, Liang, H, Liu, Y. | | 登録日 | 2014-08-27 | | 公開日 | 2015-06-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Structural and Functional Characterization of Acinetobacter baumannii Nucleoside Diphosphate Kinase
Prog.Biochem.Biophys., 42, 2015
|
|
6AGY
 
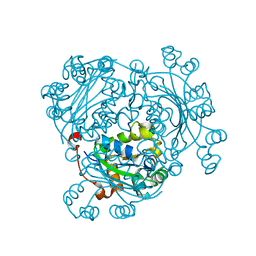 | | Aspergillus fumigatus Af293 NDK | | 分子名称: | Nucleoside diphosphate kinase | | 著者 | Hu, Y, Han, L. | | 登録日 | 2018-08-15 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Characterization of crystal structure and key residues of Aspergillus fumigatus nucleoside diphosphate kinase.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
4WBF
 
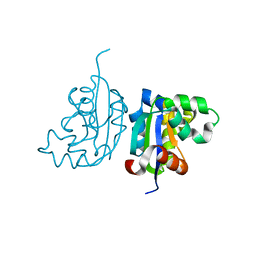 | | Acinetobacter baumannii SDF NDK | | 分子名称: | Nucleoside diphosphate kinase | | 著者 | Hu, Y, Liu, Y. | | 登録日 | 2014-09-03 | | 公開日 | 2015-06-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Structural and Functional Characterization of Acinetobacter baumannii Nucleoside Diphosphate Kinase
Prog.Biochem.Biophys., 42, 2015
|
|
2VHA
 
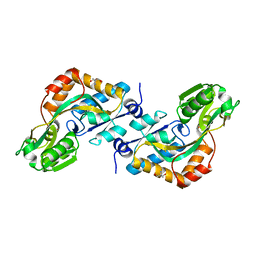 | | DEBP | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTAMIC ACID, PERIPLASMIC BINDING TRANSPORT PROTEIN | | 著者 | Hu, Y.L, Fan, C.-P, Fu, G.S, Zhu, D.Y, Jin, Q, Wang, D.-C. | | 登録日 | 2007-11-20 | | 公開日 | 2008-07-08 | | 最終更新日 | 2019-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Crystal Structure of a Glutamate/Aspartate Binding Protein Complexed with a Glutamate Molecule: Structural Basis of Ligand Specificity at Atomic Resolution.
J.Mol.Biol., 382, 2008
|
|
1NLM
 
 | | CRYSTAL STRUCTURE OF MURG:GLCNAC COMPLEX | | 分子名称: | GLYCEROL, UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | 著者 | Hu, Y, Chen, L, Ha, S, Gross, B, Falcone, B, Walker, D, Mokhtarzadeh, M, Walker, S. | | 登録日 | 2003-01-07 | | 公開日 | 2003-02-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
8H3F
 
 | | Cryo-EM Structure of the KBTBD2-CRL3-CSN complex | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (6.73 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3A
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8(removed)-CSN complex | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.51 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3Q
 
 | | Cryo-EM Structure of the CAND1-Cul3-Rbx1 complex | | 分子名称: | Cullin-3, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-09 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.76 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H35
 
 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 octameric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.41 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H36
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a dimeric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H33
 
 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 tetrameric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-07 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.86 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H34
 
 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 hexameric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-07 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.99 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3R
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8 dimeric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-09 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (6.36 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H38
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8-CSN(mutate) complex | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.25 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H37
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a tetrameric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.52 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1WOD
 
 | | CRYSTAL STRUCTURE OF MODA, A MOLYBDATE PROTEIN, COMPLEXED WITH TUNGSTATE | | 分子名称: | MODA, TUNGSTATE(VI)ION | | 著者 | Hu, Y, Rech, S, Gunsalus, R.P, Rees, D.C. | | 登録日 | 1997-06-17 | | 公開日 | 1997-12-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of the molybdate binding protein ModA.
Nat.Struct.Biol., 4, 1997
|
|
6KQX
 
 | | Crystal structure of Yijc from B. subtilis in complex with UDP | | 分子名称: | URIDINE-5'-DIPHOSPHATE, Uncharacterized UDP-glucosyltransferase YjiC | | 著者 | Hu, Y.M, Dai, L.H, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2019-08-20 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Structural dissection of unnatural ginsenoside-biosynthetic UDP-glycosyltransferase Bs-YjiC from Bacillus subtilis for substrate promiscuity.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
6KQW
 
 | | Crystal structure of Yijc from B. subtilis | | 分子名称: | CITRIC ACID, Uncharacterized UDP-glucosyltransferase YjiC | | 著者 | Hu, Y.M, Dai, L.H, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2019-08-20 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Structural dissection of unnatural ginsenoside-biosynthetic UDP-glycosyltransferase Bs-YjiC from Bacillus subtilis for substrate promiscuity.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
4TQJ
 
 | | Structural basis of specific recognition of non-reducing terminal N-acetylglucosamine by an Agrocybe aegerita lection | | 分子名称: | Lectin 2 | | 著者 | Hu, Y.L, Ren, X.M, Li, D.F, Jiang, S, Lan, X.Q, Sun, H, Wang, D.C. | | 登録日 | 2014-06-11 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis of Specific Recognition of Non-Reducing Terminal N-Acetylglucosamine by an Agrocybe aegerita Lectin.
Plos One, 10, 2015
|
|
4TQM
 
 | | Structural basis of specific recognition of non-reducing terminal N-acetylglucosamine by an Agrocybe aegerita lection | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lectin 2 | | 著者 | Hu, Y.L, Ren, X.M, Li, D.F, Jiang, S, Lan, X.Q, Sun, H, Wang, D.C. | | 登録日 | 2014-06-11 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis of Specific Recognition of Non-Reducing Terminal N-Acetylglucosamine by an Agrocybe aegerita Lectin.
Plos One, 10, 2015
|
|
4TQK
 
 | | Structural basis of specific recognition of non-reducing terminal N-acetylglucosamine by an Agrocybe aegerita lection | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lectin 2 | | 著者 | Hu, Y.L, Ren, X.M, Li, D.F, Jiang, S, Lan, X.Q, Sun, H, Wang, D.C. | | 登録日 | 2014-06-11 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis of Specific Recognition of Non-Reducing Terminal N-Acetylglucosamine by an Agrocybe aegerita Lectin.
Plos One, 10, 2015
|
|
8CMP
 
 | |
1ZLG
 
 | | Solution structure of the extracellular matrix protein anosmin-1 | | 分子名称: | Anosmin 1 | | 著者 | Hu, Y, Sun, Z, Eaton, J.T, Bouloux, P.M, Perkins, S.J. | | 登録日 | 2005-05-06 | | 公開日 | 2006-05-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION SCATTERING | | 主引用文献 | Extended and Flexible Domain Solution Structure of the Extracellular Matrix Protein Anosmin-1 by X-ray Scattering, Analytical Ultracentrifugation and Constrained Modelling.
J.Mol.Biol., 350, 2005
|
|
7UPN
 
 | |
3WE9
 
 | | The crystal structure of YisP from Bacillus subtilis subsp. subtilis strain 168 | | 分子名称: | Putative phytoene/squalene synthase YisP, TRIETHYLENE GLYCOL | | 著者 | Hu, Y, Huang, C.H, Chan, H.C, Ko, T.P, Feng, X, Oldfield, E, Guo, R.T. | | 登録日 | 2013-07-02 | | 公開日 | 2014-07-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Crystal structure of Bacillus subtilis YisP in complex with a PEG fragment
To be Published
|
|
