1PEN
 
 | | ALPHA-CONOTOXIN PNI1 | | Descriptor: | ALPHA-CONOTOXIN PNIA | | Authors: | Hu, S.-H, Gehrmann, J, Guddat, L.W, Alewood, P.F, Craik, D.J, Martin, J.L. | | Deposit date: | 1996-01-29 | | Release date: | 1997-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 A crystal structure of the neuronal acetylcholine receptor antagonist, alpha-conotoxin PnIA from Conus pennaceus.
Structure, 4, 1996
|
|
3E4H
 
 | | Crystal structure of the cyclotide varv F | | Descriptor: | Varv peptide F,Varv peptide F | | Authors: | Hu, S.H. | | Deposit date: | 2008-08-11 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Combined X-ray and NMR Analysis of the Stability of the Cyclotide Cystine Knot Fold That Underpins Its Insecticidal Activity and Potential Use as a Drug Scaffold
J.Biol.Chem., 284, 2009
|
|
3PUK
 
 | | Re-refinement of the crystal structure of Munc18-3 and Syntaxin4 N-peptide complex | | Descriptor: | Syntaxin-4 N-terminal peptide, Syntaxin-binding protein 3 | | Authors: | Hu, S.-H, Christie, M.P, Saez, N.J, Latham, C.F, Jarrott, R, Lua, L.H.L, Collins, B.M, Martin, J.L. | | Deposit date: | 2010-12-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.054 Å) | | Cite: | Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1BED
 
 | | STRUCTURE OF DISULFIDE OXIDOREDUCTASE | | Descriptor: | DSBA OXIDOREDUCTASE | | Authors: | Hu, S.-H, Martin, J.L. | | Deposit date: | 1996-09-16 | | Release date: | 1997-10-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of TcpG, the DsbA protein folding catalyst from Vibrio cholerae.
J.Mol.Biol., 268, 1997
|
|
3AJ2
 
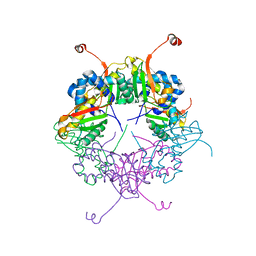 | | The structure of AxCeSD octamer (C-terminal HIS-tag) from Acetobacter xylinum | | Descriptor: | Cellulose synthase operon protein D | | Authors: | Hu, S.Q, Tajima, K, Zhou, Y, Tanaka, I, Yao, M. | | Deposit date: | 2010-05-20 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of bacterial cellulose synthase subunit D octamer with four inner passageways
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3AJ1
 
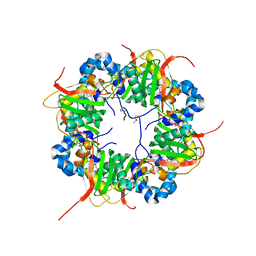 | | The structure of AxCeSD octamer (N-terminal HIS-tag) from Acetobacter xylinum | | Descriptor: | Cellulose synthase operon protein D | | Authors: | Hu, S.Q, Tajima, K, Zhou, Y, Tanaka, I, Yao, M. | | Deposit date: | 2010-05-20 | | Release date: | 2010-10-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of bacterial cellulose synthase subunit D octamer with four inner passageways
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3A8E
 
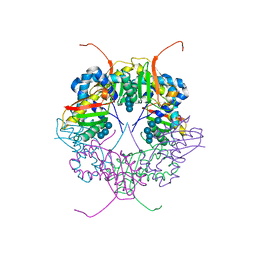 | | The structure of AxCesD octamer complexed with cellopentaose | | Descriptor: | Cellulose synthase operon protein D, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hu, S.Q, Tajima, K, Zhou, Y, Yao, M, Tanaka, I. | | Deposit date: | 2009-10-05 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of bacterial cellulose synthase subunit D octamer with four inner passageways
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1AKG
 
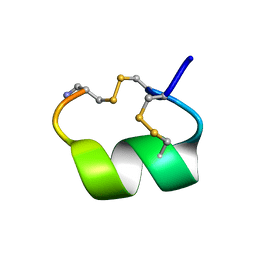 | | ALPHA-CONOTOXIN PNIB FROM CONUS PENNACEUS | | Descriptor: | ALPHA-CONOTOXIN PNIB | | Authors: | Hu, S.-H, Martin, J.L. | | Deposit date: | 1997-05-18 | | Release date: | 1998-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure at 1.1 A resolution of alpha-conotoxin PnIB: comparison with alpha-conotoxins PnIA and GI.
Biochemistry, 36, 1997
|
|
1A0M
 
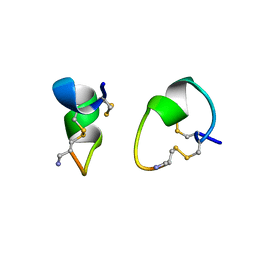 | | 1.1 ANGSTROM CRYSTAL STRUCTURE OF A-CONOTOXIN [TYR15]-EPI | | Descriptor: | ALPHA-CONOTOXIN [TYR15]-EPI | | Authors: | Hu, S.-H, Loughnan, M, Miller, R, Weeks, C.M, Blessing, R.H, Alewood, P.F, Lewis, R.J, Martin, J.L. | | Deposit date: | 1997-12-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 A resolution crystal structure of [Tyr15]EpI, a novel alpha-conotoxin from Conus episcopatus, solved by direct methods.
Biochemistry, 37, 1998
|
|
7YPW
 
 | | Lloviu cuevavirus nucleoprotein-RNA complex | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Hu, S.F, Fujita-Fujiharu, Y, Sugita, Y, Wendt, L, Muramoto, Y, Nakano, M, Hoenen, T, Noda, T. | | Deposit date: | 2022-08-04 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.0356 Å) | | Cite: | Cryoelectron microscopic structure of the nucleoprotein-RNA complex of the European filovirus, Lloviu virus.
Pnas Nexus, 2, 2023
|
|
7YR8
 
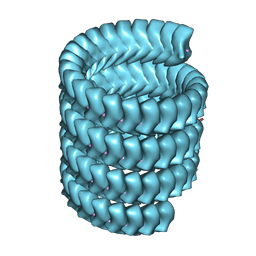 | | Lloviu cuevavirus nucleoprotein(1-450 residues)-RNA complex | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Hu, S.F, Fujita-Fujiharu, Y, Sugita, Y, Wendt, L, Muramoto, Y, Nakano, M, Hoenen, T, Noda, T. | | Deposit date: | 2022-08-09 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryoelectron microscopic structure of the nucleoprotein-RNA complex of the European filovirus, Lloviu virus.
Pnas Nexus, 2, 2023
|
|
3W94
 
 | |
3PUJ
 
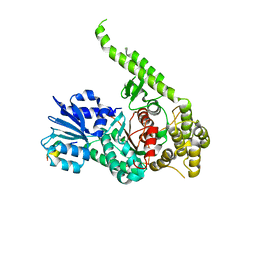 | | Crystal structure of the MUNC18-1 and SYNTAXIN4 N-Peptide complex | | Descriptor: | Syntaxin-4 N-terminal peptide, Syntaxin-binding protein 1 | | Authors: | Hu, S.-H, Christie, M.P, Saez, N.J, Latham, C.F, Jarrott, R, Lua, L.H.L, Collins, B.M, Martin, J.L. | | Deposit date: | 2010-12-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.313 Å) | | Cite: | Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6J56
 
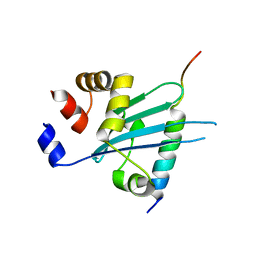 | | Crystal structure of Myosin VI CBD in complex with Tom1 MBM | | Descriptor: | Peptide from Target of Myb protein 1, Unconventional myosin-VI | | Authors: | Hu, S, Pan, L. | | Deposit date: | 2019-01-10 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structure of Myosin VI/Tom1 complex reveals a cargo recognition mode of Myosin VI for tethering.
Nat Commun, 10, 2019
|
|
3WLW
 
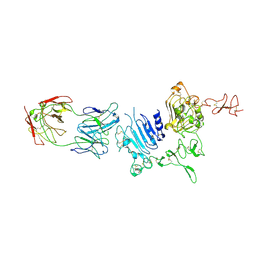 | | Molecular Architecture of the ErbB2 Extracellular Domain Homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody H Chain, ... | | Authors: | Hu, S, Lou, Z.Y, Guo, Y.J. | | Deposit date: | 2013-11-15 | | Release date: | 2015-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.088 Å) | | Cite: | Molecular architecture of the ErbB2 extracellular domain homodimer.
Oncotarget, 6, 2015
|
|
5ZVL
 
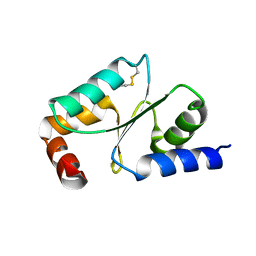 | | Crystal Structure of Wheat Glutarredoxin | | Descriptor: | Glutaredoxin | | Authors: | Hu, S.Q, Sun, X.M, Chen, M.R. | | Deposit date: | 2018-05-11 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.963 Å) | | Cite: | Crystal Structure of Wheat Glutaredoxin and Its Application in Improving the Processing Quality of Flour.
J. Agric. Food Chem., 66, 2018
|
|
8X6B
 
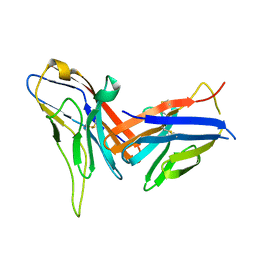 | | Crystal structure of immune receptor PVRIG in complex with ligand Nectin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nectin-2, Transmembrane protein PVRIG | | Authors: | Hu, S.T, Han, P, Wang, H, Qi, J.X. | | Deposit date: | 2023-11-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the immune recognition and selectivity of the immune receptor PVRIG for ligand Nectin-2.
Structure, 32, 2024
|
|
3WD5
 
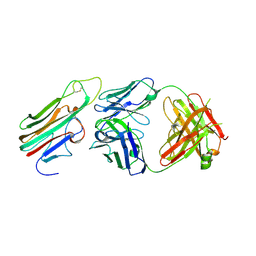 | | Crystal structure of TNFalpha in complex with Adalimumab Fab fragment | | Descriptor: | Adalimumab Heavy Chain, Adalimumab Light Chain, Tumor necrosis factor | | Authors: | Hu, S, Liang, S.Y, Guo, Y.J, Lou, Z.Y. | | Deposit date: | 2013-06-06 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Comparison of the inhibition mechanisms of adalimumab and infliximab in treating tumor necrosis factor alpha-associated diseases from a molecular view
J.Biol.Chem., 288, 2013
|
|
7F5P
 
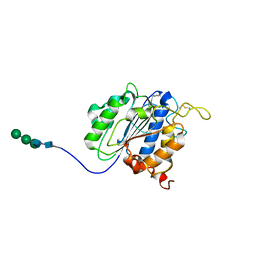 | | The crystal structure of VyPAL2-C214A, a dead mutant of VyPAL2 peptide asparaginyl ligase in form I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Peptide Asparaginyl Ligases, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
7F5Q
 
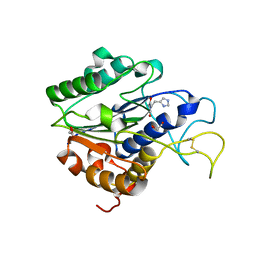 | | The crystal structure of VyPAL2 peptide asparaginyl ligase in its active enzyme form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
7FA0
 
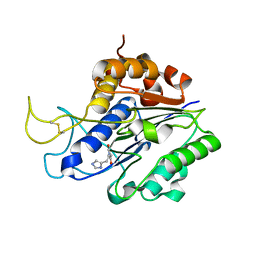 | | The crystal structure of VyPAL2-C214A, a dead mutant of VyPAL2 peptide asparaginyl ligase in form II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Peptide Asparaginyl Ligases, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-07-05 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
7F5J
 
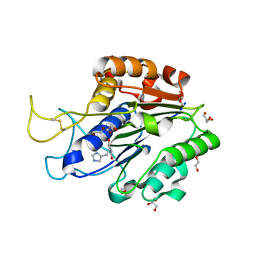 | | The crystal structure of VyPAL2-I244V, a more efficient mutant of VyPAL2 peptide asparaginyl ligase in its active enzyme form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
3JBH
 
 | | TWO HEAVY MEROMYOSIN INTERACTING-HEADS MOTIFS FLEXIBLE DOCKED INTO TARANTULA THICK FILAMENT 3D-MAP ALLOWS IN DEPTH STUDY OF INTRA- AND INTERMOLECULAR INTERACTIONS | | Descriptor: | MYOSIN 2 ESSENTIAL LIGHT CHAIN STRIATED MUSCLE, MYOSIN 2 HEAVY CHAIN STRIATED MUSCLE, MYOSIN 2 REGULATORY LIGHT CHAIN STRIATED MUSCLE | | Authors: | Alamo, L, Qi, D, Wriggers, W, Pinto, A, Zhu, J, Bilbao, A, Gillilan, R.E, Hu, S, Padron, R. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis.
J. Mol. Biol., 428, 2016
|
|
3JSD
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
3BXQ
 
 | | The structure of a mutant insulin uncouples receptor binding from protein allostery. An electrostatic block to the TR transition | | Descriptor: | ZINC ION, insulin A chain, insulin B chain | | Authors: | Wan, Z.L, Huang, K, Hu, S.Q, Whittaker, J, Weiss, M.A. | | Deposit date: | 2008-01-14 | | Release date: | 2008-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of a mutant insulin uncouples receptor binding from protein allostery. An electrostatic block to the TR transition.
J.Biol.Chem., 283, 2008
|
|
