1W9N
 
 | | Isolation and characterization of epilancin 15X, a novel antibiotic from a clinical strain of Staphylococcus epidermidis | | Descriptor: | EPILANCIN 15X | | Authors: | Ekkelenkamp, M, Hanssen, M.G.M, Hsu, S.-T.D, de Jong, A, Milatovic, D, Verhoef, J, van Nuland, N.A.J. | | Deposit date: | 2004-10-14 | | Release date: | 2005-04-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Isolation and structural characterization of epilancin 15X, a novel lantibiotic from a clinical strain of Staphylococcus epidermidis.
FEBS Lett., 579, 2005
|
|
1R0M
 
 | | Structure of Deinococcus radiodurans N-acylamino acid racemase at 1.3 : insights into a flexible binding pocket and evolution of enzymatic activity | | Descriptor: | N-acylamino acid racemase | | Authors: | Wang, W.-C, Chiu, W.-C, Hsu, S.-K, Wu, C.-L, Chen, C.-Y, Liu, J.-S, Hsu, W.-H. | | Deposit date: | 2003-09-22 | | Release date: | 2004-09-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans
J.Mol.Biol., 342, 2004
|
|
8W68
 
 | | Crystal structure of Q9PR55 at pH 6.0 (use NMR model) | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-08-28 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8IWA
 
 | | Crystal structure of Q9PR55 at pH 6.5 | | Descriptor: | SULFATE ION, Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
2X6M
 
 | | Structure of a single domain camelid antibody fragment in complex with a C-terminal peptide of alpha-synuclein | | Descriptor: | ALPHA-SYNUCLEIN PEPTIDE, HEAVY CHAIN VARIABLE DOMAIN FROM DROMEDARY | | Authors: | DeGenst, E, Guilliams, T, Wellens, J, O'Day, E.M, Waudby, C.A, Meehan, S, Dumoulin, M, Hsu, S.-T.D, Cremades, N, Verschueren, K.H.G, Pardon, E, Wyns, L, Steyaert, J, Christodoulou, J, Dobson, C.M. | | Deposit date: | 2010-02-18 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure and Properties of a Complex of Alpha-Synuclein and a Single-Domain Camelid Antibody.
J.Mol.Biol., 402, 2010
|
|
8IWC
 
 | | Crystal structure of Q9PR55 at pH 6.0 | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8IWB
 
 | | Crystal structure of Q9PR55 at pH 7.5 | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
7Y39
 
 | | Ubiquitin-like domain of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1 | | Authors: | Lai, C.H, Ko, K.T, Fan, P.J, Yu, T.A, Chang, C.F, Draczkowski, P, Hsu, S.T.D. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Insight into ZFAND1 and p97 Interaction
To Be Published
|
|
7Y7L
 
 | | Solution structure of zinc finger domain 2 of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1, ZINC ION | | Authors: | Fang, P.J, Lai, C.H, Ko, K.T, Chang, C.F, Hsu, S.T.D. | | Deposit date: | 2022-06-22 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of p97 recognition by human ZFAND1
To Be Published
|
|
7YAB
 
 | | Solution structure of zinc finger domain 1 of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1, ZINC ION | | Authors: | Fang, P.J, Lai, C.H, Ko, K.T, Chang, C.F, Hsu, S.T.D. | | Deposit date: | 2022-06-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of p97 recognition by human ZFAND1
To Be Published
|
|
1XS2
 
 | | Structural Basis for Catalytic Racemization and Substrate Specificity of an N-Acylamino Acid Racemase Homologue from Deinococcus radiodurans | | Descriptor: | MAGNESIUM ION, N-Acylamino Acid Racemase | | Authors: | Wang, W.-C, Chiu, W.-C, Hsu, S.-K, Wu, C.-L, Chen, C.-Y, Liu, J.-S, Hsu, W.-H. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans
J.Mol.Biol., 342, 2004
|
|
1XPY
 
 | | Structural Basis for Catalytic Racemization and Substrate Specificity of an N-Acylamino Acid Racemase Homologue from Deinococcus radiodurans | | Descriptor: | MAGNESIUM ION, N-acylamino acid racemase, N~2~-ACETYL-L-GLUTAMINE | | Authors: | Wang, W.-C, Chiu, W.-C, Hsu, S.-K, Wu, C.-L, Chen, C.-Y, Liu, J.-S, Hsu, W.-H. | | Deposit date: | 2004-10-10 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans
J.Mol.Biol., 342, 2004
|
|
7YJW
 
 | |
2BUN
 
 | | Solution structure of the BLUF domain of AppA 5-125 | | Descriptor: | APPA, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Grinstead, J.S, Hsu, S.-T, Laan, W, Bonvin, A.M.J.J, Hellingwerf, K.J, Boelens, R, Kaptein, R. | | Deposit date: | 2005-06-15 | | Release date: | 2005-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the AppA BLUF domain: insight into the mechanism of light-induced signaling.
Chembiochem, 7, 2006
|
|
3WRE
 
 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
3WRG
 
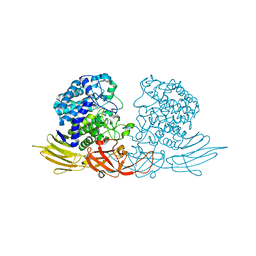 | | The complex structure of HypBA1 with L-arabinose | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION, beta-L-arabinofuranose | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
3WRF
 
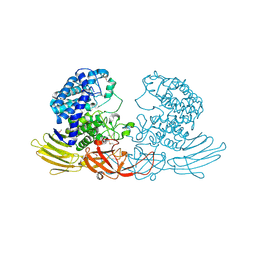 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
1L3M
 
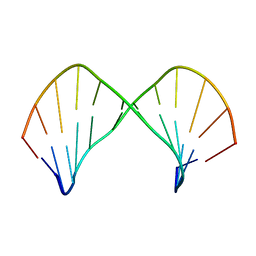 | | The Solution Structure of [d(CGC)r(amamam)d(TTTGCG)]2 | | Descriptor: | 5'-D(*CP*GP*C)-R(P*(A39)P*(A39)P*(A39))-D(P*TP*TP*TP*GP*CP*G)-3' | | Authors: | Tsao, Y.P, Wang, L.Y, Hsu, S.T, Jain, M.L, Chou, S.H, Huang, W.C, Cheng, J.W. | | Deposit date: | 2002-02-28 | | Release date: | 2002-04-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of [d(CGC)r(amamam)d(TTTGCG)]2.
J.Biomol.NMR, 21, 2001
|
|
2LRV
 
 | |
2LRM
 
 | |
8HW9
 
 | | Solution structure of ubiquitin-like domain (UBL) of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1 | | Authors: | Lai, C.H, Ko, K.T, Fan, P.J, Yu, T.A, Chang, C.F, Hsu, S.T.D. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ubiquitin-like domain (UBL) of human ZFAND1
To Be Published
|
|
1SIY
 
 | | NMR structure of mung bean non-specific lipid transfer protein 1 | | Descriptor: | Nonspecific lipid-transfer protein 1 | | Authors: | Lin, K.F, Liu, Y.N, Hsu, S.T.D, Samuel, D, Cheng, C.S, Bonvin, A.M.J.J, Lyu, P.C. | | Deposit date: | 2004-03-02 | | Release date: | 2005-04-05 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Characterization and Structural Analyses of Nonspecific Lipid Transfer Protein 1 from Mung Bean
Biochemistry, 44, 2005
|
|
6LX0
 
 | |
6AHW
 
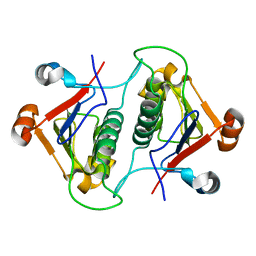 | |
5ZYO
 
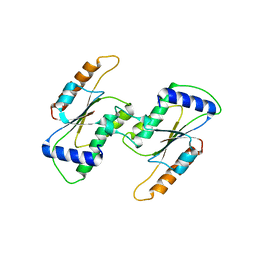 | |
