5WFX
 
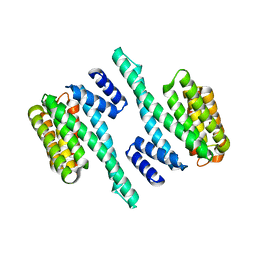 | |
5WFU
 
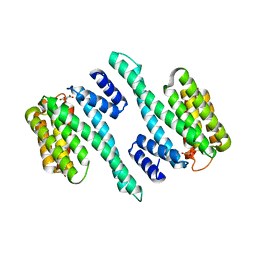 | |
5VJI
 
 | | Crystal structure of the CLOCK Transcription Domain Exon19 in Complex with a Repressor | | Descriptor: | CLOCK-interacting pacemaker, Circadian locomoter output cycles protein kaput | | Authors: | Hou, Z, Su, L, Pei, J, Grishin, N.V, Zhang, H. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-07 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the CLOCK Transactivation Domain Exon19 in Complex with a Repressor.
Structure, 25, 2017
|
|
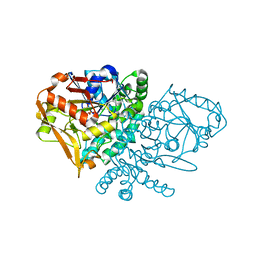
5VJX
 
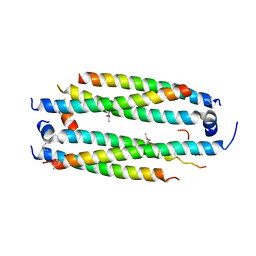 | | Crystal structure of the CLOCK Transcription Domain Exon19 in Complex with a Repressor | | Descriptor: | CLOCK-interacting pacemaker, Circadian locomoter output cycles protein kaput | | Authors: | Hou, Z, Su, L, Pei, J, Grishin, N.V, Zhang, H. | | Deposit date: | 2017-04-20 | | Release date: | 2017-12-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Crystal Structure of the CLOCK Transactivation Domain Exon19 in Complex with a Repressor.
Structure, 25, 2017
|
|
1KKF
 
 | | Complex of E. coli Adenylosuccinate Synthetase with IMP, Hadacidin, Pyrophosphate, and Mg | | Descriptor: | Adenylosuccinate Synthetase, DIPHOSPHATE, HADACIDIN, ... | | Authors: | Hou, Z, Wang, W, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2001-12-07 | | Release date: | 2002-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | IMP Alone Organizes the Active Site of Adenylosuccinate Synthetase from Escherichia coli.
J.Biol.Chem., 277, 2002
|
|
1KJX
 
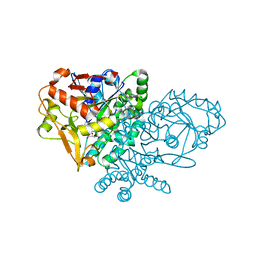 | | IMP Complex of E. Coli Adenylosuccinate Synthetase | | Descriptor: | Adenylosuccinate Synthetase, INOSINIC ACID | | Authors: | Hou, Z, Wang, W, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | IMP Alone Organizes the Active Site of Adenylosuccinate Synthetase from Escherichia coli.
J.Biol.Chem., 277, 2002
|
|
1KKB
 
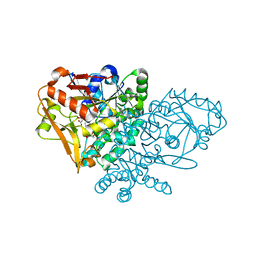 | | Complex of Escherichia coli Adenylosuccinate Synthetase with IMP and Hadacidin | | Descriptor: | Adenylosuccinate Synthetase, HADACIDIN, INOSINIC ACID | | Authors: | Hou, Z, Wang, W, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2001-12-06 | | Release date: | 2002-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | IMP Alone Organizes the Active Site of Adenylosuccinate Synthetase from Escherichia coli.
J.Biol.Chem., 277, 2002
|
|
1CIB
 
 | | STRUCTURE OF ADENYLOSUCCINATE SYNTHETASE FROM E. COLI COMPLEXED WITH GDP, IMP, HADACIDIN, AND NO3 | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE, HADACIDIN, ... | | Authors: | Hou, Z, Cashel, M, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1999-03-31 | | Release date: | 2000-04-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effectors of the stringent response target the active site of Escherichia coli adenylosuccinate synthetase.
J.Biol.Chem., 274, 1999
|
|
1ZHI
 
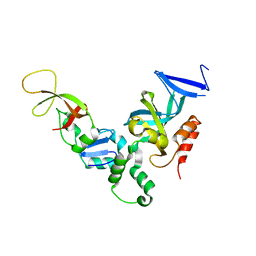 | | Complex of the S. cerevisiae Orc1 and Sir1 interacting domains | | Descriptor: | Origin recognition complex subunit 1, Regulatory protein SIR1 | | Authors: | Hou, Z, Bernstein, D.A, Fox, C.A, Keck, J.L. | | Deposit date: | 2005-04-25 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the Sir1-origin recognition complex interaction in transcriptional silencing.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1Z1A
 
 | |
3VAY
 
 | | Crystal structure of 2-Haloacid Dehalogenase from Pseudomonas syringae pv. Tomato DC3000 | | Descriptor: | HAD-superfamily hydrolase, IODIDE ION, MAGNESIUM ION | | Authors: | Hou, Z, Zhang, H, Li, M, Chang, W. | | Deposit date: | 2011-12-30 | | Release date: | 2013-01-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structure of 2-haloacid dehalogenase from Pseudomonas syringae pv. tomato DC3000
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1CH8
 
 | | STRUCTURE OF ADENYLOSUCCINATE SYNTHETASE FROM E. COLI COMPLEXED WITH A STRINGENT EFFECTOR, PPG2':3'P | | Descriptor: | GUANOSINE 5'-DIPHOSPHATE 2':3'-CYCLIC MONOPHOSPHATE, HADACIDIN, INOSINIC ACID, ... | | Authors: | Hou, Z, Cashel, M, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1999-03-31 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Effectors of the stringent response target the active site of Escherichia coli adenylosuccinate synthetase.
J.Biol.Chem., 274, 1999
|
|
8KHU
 
 | | Hepatitis B virus core protein Y132A mutant in complex with THPP derivatives 48 | | Descriptor: | (6~{S},7~{R})-6,7-dimethyl-3-(2-oxidanylidenepyrrolidin-1-yl)-~{N}-[3,4,5-tris(fluoranyl)phenyl]-6,7-dihydro-4~{H}-pyrazolo[1,5-a]pyrazine-5-carboxamide, Capsid protein, GLYCEROL, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2023-08-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4,5,6,7-Tetrahydropyrazolo[1.5-a]pyrizine Derivatives as Core Protein Allosteric Modulators (CpAMs) for the Inhibition of Hepatitis B Virus.
J.Med.Chem., 66, 2023
|
|
2L5A
 
 | | Structural basis for recognition of centromere specific histone H3 variant by nonhistone Scm3 | | Descriptor: | Histone H3-like centromeric protein CSE4, Protein SCM3, Histone H4 | | Authors: | Zhou, Z, Feng, H, Zhou, B, Ghirlando, R, Hu, K, Zwolak, A, Jenkins, L, Xiao, H, Tjandra, N, Wu, C, Bai, Y. | | Deposit date: | 2010-10-28 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3.
Nature, 472, 2011
|
|
7XGL
 
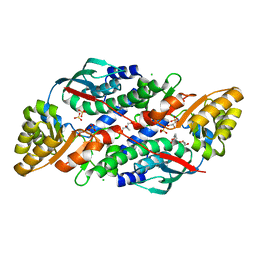 | | Quinolinate Phosphoribosyl Transferase (QAPRTase) from Streptomyces pyridomyceticus NRRL B-2517 in Apo form | | Descriptor: | CHLORIDE ION, GLYCEROL, Quinolinate Phosphoribosyl Transferase, ... | | Authors: | Zhou, Z, Yang, X, Huang, T, Wang, X, Liang, R, Zheng, J, Dai, S, Lin, S, Deng, Z. | | Deposit date: | 2022-04-05 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Bifunctional NadC Homologue PyrZ Catalyzes Nicotinic Acid Formation in Pyridomycin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
7XGM
 
 | | Quinolinate Phosphoribosyl Transferase (QAPRTase) from Streptomyces pyridomyceticus NRRL B-2517 in complex with Quinolinic Acid (QA) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, QUINOLINIC ACID, ... | | Authors: | Zhou, Z, Yang, X, Huang, T, Wang, X, Liang, R, Zheng, J, Dai, S, Lin, S, Deng, Z. | | Deposit date: | 2022-04-05 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bifunctional NadC Homologue PyrZ Catalyzes Nicotinic Acid Formation in Pyridomycin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
7XGN
 
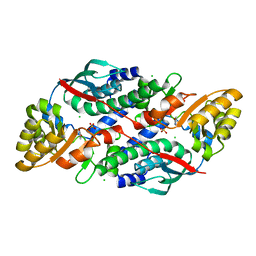 | | Quinolinate Phosphoribosyl Transferase (QAPRTase) from Streptomyces pyridomyceticus NRRL B-2517 in complex with Nicotinic Acid (NA) | | Descriptor: | CHLORIDE ION, NICOTINIC ACID, Quinolinate Phosphoribosyl Transferase, ... | | Authors: | Zhou, Z, Yang, X, Huang, T, Wang, X, Liang, R, Zheng, J, Dai, S, Lin, S, Deng, Z. | | Deposit date: | 2022-04-05 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bifunctional NadC Homologue PyrZ Catalyzes Nicotinic Acid Formation in Pyridomycin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
2JUJ
 
 | |
2QJU
 
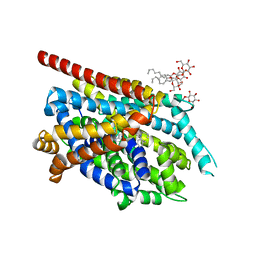 | | Crystal Structure of an NSS Homolog with Bound Antidepressant | | Descriptor: | 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, CHLORIDE ION, LEUCINE, ... | | Authors: | Zhou, Z, Karpowich, N.K, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2007-07-09 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake.
Science, 317, 2007
|
|
2RPI
 
 | |
3TB3
 
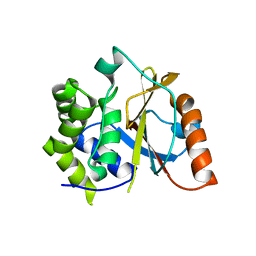 | | Crystal structure of the UCH domain of UCH-L5 with 6 residues deleted | | Descriptor: | CALCIUM ION, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Zhou, Z.R, Zha, M, Zhou, J, Hu, H.Y. | | Deposit date: | 2011-08-05 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Length of the active-site crossover loop defines the substrate specificity of ubiquitin C-terminal hydrolases for ubiquitin chains.
Biochem.J., 441, 2012
|
|
1SV2
 
 | | Crystal Structure of Peptide Deformylase from Leptospira Interrogans (LiPDF) at pH7.5 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, FORMIC ACID, Peptide deformylase, ... | | Authors: | Zhou, Z, Song, X, Li, Y, Gong, W. | | Deposit date: | 2004-03-27 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel conformational states of peptide deformylase from pathogenic bacterium Leptospira interrogans: implications for population shift
J.Biol.Chem., 280, 2005
|
|
1Y6H
 
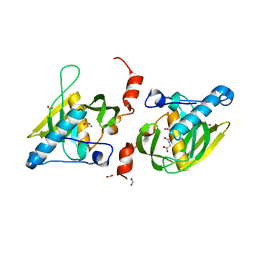 | | Crystal structure of LIPDF | | Descriptor: | FORMIC ACID, GLYCINE, Peptide deformylase, ... | | Authors: | Zhou, Z, Song, X, Li, Y, Gong, W. | | Deposit date: | 2004-12-06 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique structural characteristics of peptide deformylase from pathogenic bacterium Leptospira interrogans
J.Mol.Biol., 339, 2004
|
|
7LWY
 
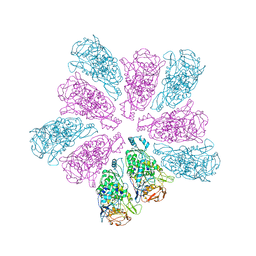 | | TVV viral capsid protein | | Descriptor: | Capsid protein | | Authors: | Zhou, Z.H, Stevens, A.W, Cui, Y.X, Johnson, P.J, Muratore, K.A. | | Deposit date: | 2021-03-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Atomic Structure of the Trichomonas vaginalis Double-Stranded RNA Virus 2.
Mbio, 12, 2021
|
|
5WTW
 
 | |
