7WIH
 
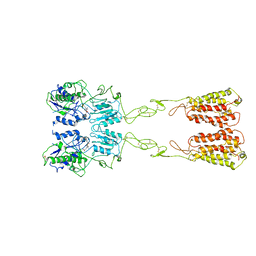 | | Cryo-EM structure of LY2794193-bound mGlu3 | | Descriptor: | (1S,2S,4S,5R,6S)-2-amino-4-[(3-methoxybenzene-1-carbonyl)amino]bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
7WI8
 
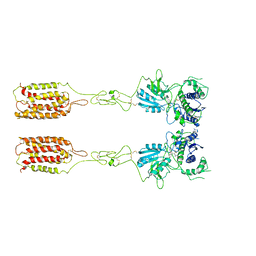 | | Cryo-EM structure of inactive mGlu3 bound to LY341495 | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
6LDU
 
 | |
6LE7
 
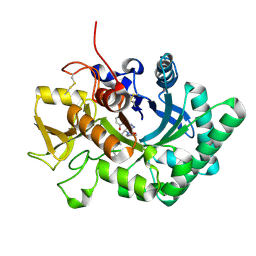 | | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 2 | | Descriptor: | (4R)-3-(2-hydroxyphenyl)-4-(3-methoxy-4-propoxy-phenyl)-5-(pyridin-3-ylmethyl)-1,4-dihydropyrrolo[3,4-c]pyrazol-6-one, Probable endochitinase | | Authors: | Chen, Q, Yang, Q, Zhou, Y. | | Deposit date: | 2019-11-24 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85753441 Å) | | Cite: | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 2
To Be Published
|
|
1C1J
 
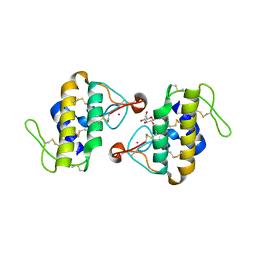 | | STRUCTURE OF CADMIUM-SUBSTITUTED PHOSPHOLIPASE A2 FROM AGKISTRONDON HALYS PALLAS AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | BASIC PHOSPHOLIPASE A2, CADMIUM ION, octyl beta-D-glucopyranoside | | Authors: | Zhang, H.-l, Zhang, Y.-q, Song, S.-y, Zhou, Y, Lin, Z.-j. | | Deposit date: | 1999-07-22 | | Release date: | 2002-07-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Cadmium-substituted Phospholipase A2 from Agkistrodon halys
Pallas at 2.8 Angstroms Resolution
Protein Pept.Lett., 6, 1999
|
|
5Y2B
 
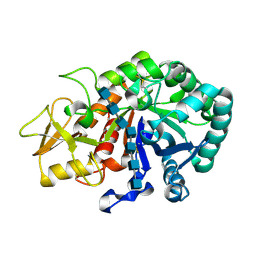 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with HEPTA-N-ACETYLCHITOOCTAOSE (NAG)7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, insect group II chitinase | | Authors: | Chen, W, Qu, M.B, Zhou, Y, Yang, Q. | | Deposit date: | 2017-07-24 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of group II chitinase (ChtII) catalysis completes the puzzle of chitin hydrolysis in insects
J. Biol. Chem., 293, 2018
|
|
5Y2C
 
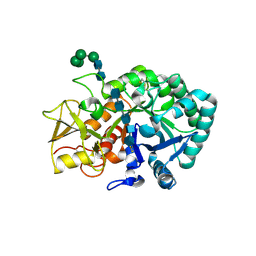 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 2 E2180L mutant in complex with PENTA-N-ACETYLCHITOOCTAOSE (NAG)5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, insect group II chitinase | | Authors: | Chen, W, Qu, M.B, Zhou, Y, Yang, Q. | | Deposit date: | 2017-07-24 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural analysis of group II chitinase (ChtII) catalysis completes the puzzle of chitin hydrolysis in insects
J. Biol. Chem., 293, 2018
|
|
7VRG
 
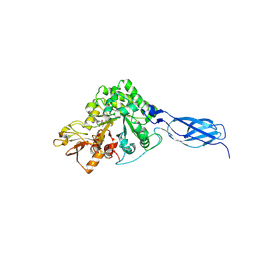 | | Crystal structure of chitinase-h from O. furnacalis in complex with Lynamicin B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, ... | | Authors: | Lu, Q, Liu, T, Zhou, Y, Yang, Q. | | Deposit date: | 2021-10-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lynamicin B is a Potential Pesticide by Acting as a Lepidoptera-Exclusive Chitinase Inhibitor.
J.Agric.Food Chem., 69, 2021
|
|
3BGK
 
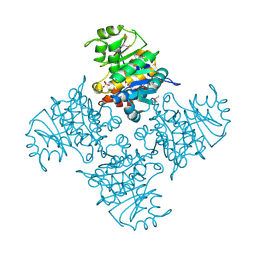 | |
7W0V
 
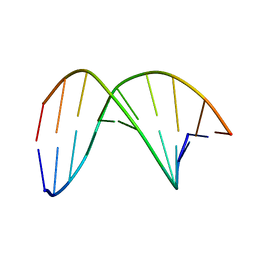 | | C4'-SCF3-DT modifeid DNA-DNA duplex | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*(DSW)P*AP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*GP*G)-3') | | Authors: | Li, Q, Trajkovski, M, Fan, C, Chen, J, Zhou, Y, Lu, K, Li, H, Su, X, Xi, Z, Plavec, J, Zhou, C. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 4'-SCF 3 -Labeling Constitutes a Sensitive 19 F NMR Probe for Characterization of Interactions in the Minor Groove of DNA.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
2GIZ
 
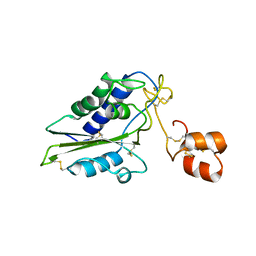 | | Structural and functional analysis of Natrin, a member of crisp-3 family blocks a variety of ion channels | | Descriptor: | Natrin-1 | | Authors: | Jiang, T, Wang, F, Li, H, Yin, C, Zhou, Y, Shu, Y, Qi, Z, Lin, Z. | | Deposit date: | 2006-03-30 | | Release date: | 2006-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and functional analysis of natrin, a venom protein that targets various ion channels
Biochem.Biophys.Res.Commun., 351, 2006
|
|
7VUH
 
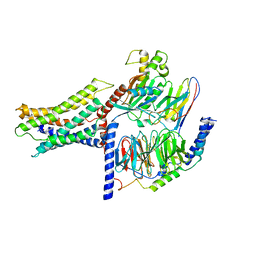 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUJ
 
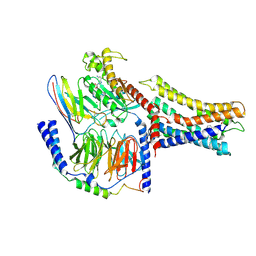 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUI
 
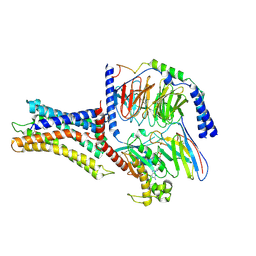 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Chimera of Endo-1,4-beta-xylanase and Probable G-protein coupled receptor 139, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUG
 
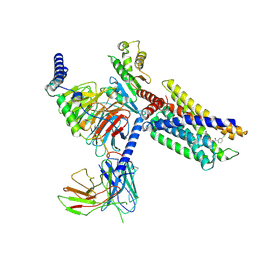 | | Cryo-EM structure of a class A orphan GPCR in complex with Gi | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Chimera of Endo-1,4-beta-xylanase and Probable G-protein coupled receptor 139, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
6JAW
 
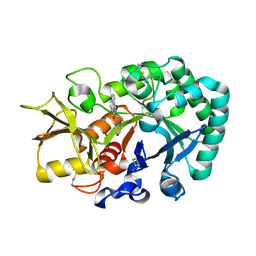 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with a napthalimide derivative | | Descriptor: | 2-[3-(morpholin-4-yl)propyl]-1H-benzo[de]isoquinoline-1,3(2H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, Group II chitinase | | Authors: | Chen, W, Zhou, Y, Yang, Q. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
6JAV
 
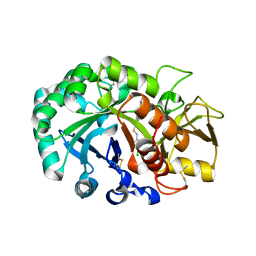 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with a piperidine-thienopyridine derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-chlorophenyl)methyl]sulfanyl}-7-methyl-N-(prop-2-en-1-yl)-7,8-dihydropyrido[4',3':4,5]thieno[2,3-d]pyrimidin-4-amine, Group II chitinase | | Authors: | Chen, W, Zhou, Y, Yang, Q. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.437 Å) | | Cite: | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
6JAY
 
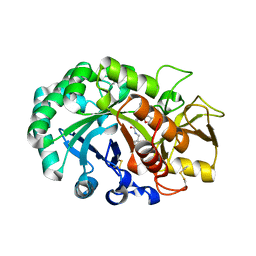 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with a dipyrido-pyrimidine derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-1-[(furan-2-yl)methyl]-5-oxo-3-({[(2S)-oxolan-2-yl]methyl}carbamoyl)-5H-dipyrido[1,2-a:2',3'-d]pyrimidin-1-ium, Group II chitinase | | Authors: | Chen, W, Zhou, Y, Yang, Q. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
6JAX
 
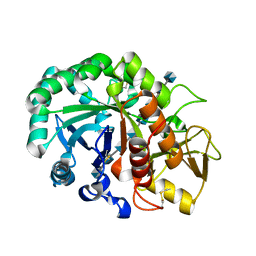 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with chitooctaose [(GlcN)8] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Group II chitinase | | Authors: | Chen, W, Zhou, Y, Yang, Q. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
2J6P
 
 | | STRUCTURE OF AS-SB REDUCTASE FROM LEISHMANIA MAJOR | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SB(V)-AS(V) REDUCTASE, ... | | Authors: | Bisacchi, D, Zhou, Y, Rosen, B.P, Mukhopadhyay, R, Bordo, D. | | Deposit date: | 2006-10-02 | | Release date: | 2007-10-02 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural characterization of the As/Sb reductase LmACR2 from Leishmania major.
J. Mol. Biol., 386, 2009
|
|
6IW7
 
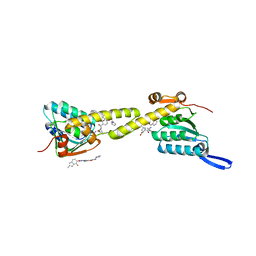 | | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease | | Descriptor: | (2S)-2-benzamido-4-methyl-pentanoic acid, 4-[(4-fluoro-2-methyl-3H-indol-5-yl)oxy]-6-methoxy-7-[3-(pyrrolidin-1-yl)propoxy]quinazoline, ATP-dependent Clp protease proteolytic subunit 1, ... | | Authors: | Bao, R, Luo, Y.F, Zhu, Y.B, Yang, Y, Zhou, Y.Z. | | Deposit date: | 2018-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.692121 Å) | | Cite: | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease
To Be Published
|
|
2GCQ
 
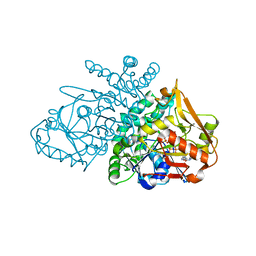 | |
3IS1
 
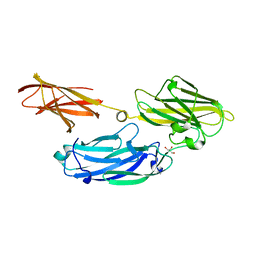 | | Crystal structure of functional region of UafA from Staphylococcus saprophyticus in C2 form at 2.45 angstrom resolution | | Descriptor: | GLYCEROL, Uro-adherence factor A | | Authors: | Tanaka, Y, Matsuoka, E, Shouji, Y, Kuroda, M, Tanaka, I, Yao, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the functional region of Uro-adherence factor A from Staphylococcus saprophyticus reveals participation of the B domain in ligand binding
Protein Sci., 20, 2011
|
|
3IS0
 
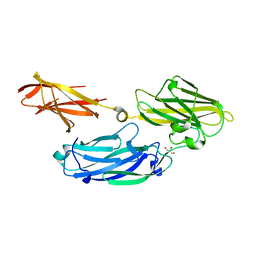 | | Crystal structure of functional region of UafA from Staphylococcus saprophyticus in the presence of cholesterol | | Descriptor: | GLYCEROL, Uro-adherence factor A | | Authors: | Tanaka, Y, Shouji, Y, Matsuoka, E, Kuroda, M, Tanaka, I, Yao, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Functional Region of UafA from Staphylococcus saprophyticus
To be Published
|
|
3IRP
 
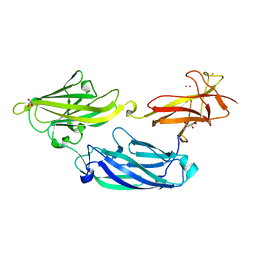 | | Crystal structure of functional region of UafA from Staphylococcus saprophyticus at 1.50 angstrom resolution | | Descriptor: | GLYCEROL, POTASSIUM ION, Uro-adherence factor A | | Authors: | Tanaka, Y, Shouji, Y, Matsuoka, E, Kuroda, M, Tanaka, I, Yao, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the functional region of Uro-adherence factor A from Staphylococcus saprophyticus reveals participation of the B domain in ligand binding
Protein Sci., 20, 2011
|
|
