2ZUT
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, GLYCEROL, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
2ZUS
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase | | Descriptor: | Lacto-N-biose phosphorylase, MAGNESIUM ION | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
2ZUV
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GlcNAc, Ethylene glycol, and nitrate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
2ZUU
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
1WQL
 
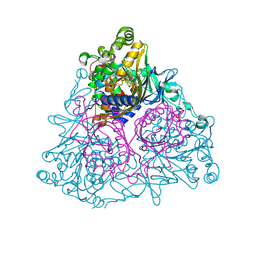 | | Cumene dioxygenase (cumA1A2) from Pseudomonas fluorescens IP01 | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, OXYGEN MOLECULE, ... | | Authors: | Dong, X, Fushinobu, S, Fukuda, E, Terada, T, Nakamura, S, Shimizu, K, Nojiri, H, Omori, T, Shoun, H, Wakagi, T. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Terminal Oxygenase Component of Cumene Dioxygenase from Pseudomonas fluorescens IP01
J.BACTERIOL., 187, 2005
|
|
1WD3
 
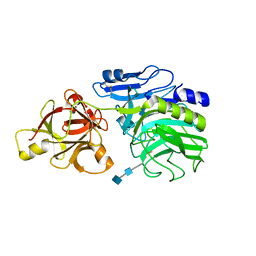 | | Crystal structure of arabinofuranosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-arabinofuranosidase B | | Authors: | Miyanaga, A, Koseki, T, Matsuzawa, H, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2004-05-11 | | Release date: | 2004-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a family 54 alpha-L-arabinofuranosidase reveals a novel carbohydrate-binding module that can bind arabinose
J.Biol.Chem., 279, 2004
|
|
1WD4
 
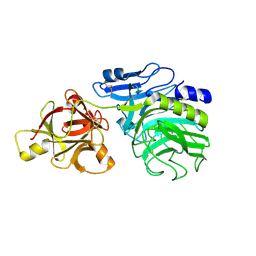 | | Crystal structure of arabinofuranosidase complexed with arabinose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-arabinofuranose, alpha-L-arabinofuranosidase B | | Authors: | Miyanaga, A, Koseki, T, Matsuzawa, H, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2004-05-11 | | Release date: | 2004-09-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of a family 54 alpha-L-arabinofuranosidase reveals a novel carbohydrate-binding module that can bind arabinose
J.Biol.Chem., 279, 2004
|
|
1XQD
 
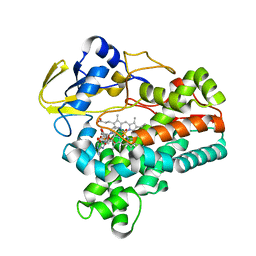 | | Crystal structure of P450NOR complexed with 3-pyridinealdehyde adenine dinucleotide | | Descriptor: | CYTOCHROME P450 55A1, NICOTINIC ACID ADENINE DINUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oshima, R, Fushinobu, S, Takaya, N, Su, F, Wakagi, T, Shoun, H. | | Deposit date: | 2004-10-12 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for direct hydride transfer from NADH to cytochrome P450nor
J.Mol.Biol., 342, 2004
|
|
1ULW
 
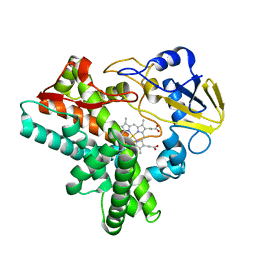 | | Crystal structure of P450nor Ser73Gly/Ser75Gly mutant | | Descriptor: | Cytochrome P450 55A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oshima, R, Fushinobu, S, Su, F, Li, Z, Takaya, N, Shoun, H. | | Deposit date: | 2003-09-16 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for direct hydride transfer from NADH to cytochrome P450nor
J.Mol.Biol., 342, 2004
|
|
1UMG
 
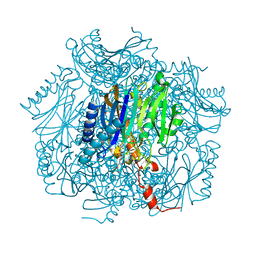 | | Crystal structure of fructose-1,6-bisphosphatase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), 385aa long conserved hypothetical protein, ... | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2003-09-30 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first crystal structure of the novel class of fructose-1,6-bisphosphatase present in thermophilic archaea.
Structure, 12, 2004
|
|
3ACG
 
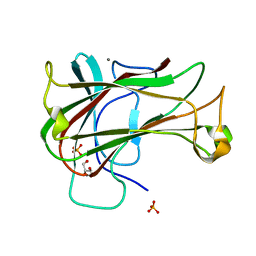 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in complex with cellobiose | | Descriptor: | Beta-1,4-endoglucanase, CALCIUM ION, GLYCEROL, ... | | Authors: | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
1V7W
 
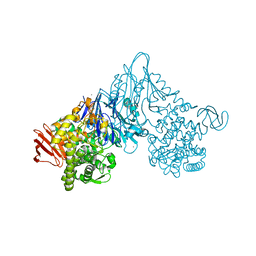 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
3ACH
 
 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in complex with cellotetraose | | Descriptor: | Beta-1,4-endoglucanase, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
2JTC
 
 | | 3D structure and backbone dynamics of SPE B | | Descriptor: | Streptopain | | Authors: | Chuang, W, Wang, C, Houng, H, Chen, C, Wang, P. | | Deposit date: | 2007-07-26 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of streptopain: insight into diverse substrate specificity.
J.Biol.Chem., 284, 2009
|
|
3ACI
 
 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in complex with cellopentaose | | Descriptor: | Beta-1,4-endoglucanase, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
3ACF
 
 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in a ligand-free form | | Descriptor: | Beta-1,4-endoglucanase, CALCIUM ION, SULFATE ION | | Authors: | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
1WU5
 
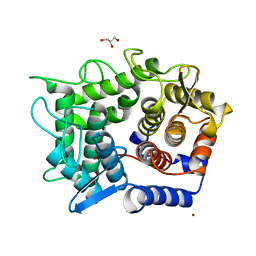 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase complexed with xylose | | Descriptor: | GLYCEROL, NICKEL (II) ION, beta-D-xylopyranose, ... | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Specificity of the Reducing End Xylose-releasing Exo-oligoxylanase from Bacillus halodurans C-125
J.Biol.Chem., 280, 2005
|
|
1WU4
 
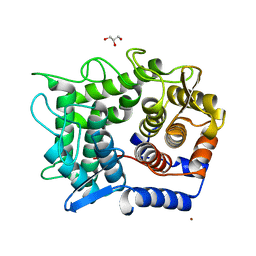 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase | | Descriptor: | GLYCEROL, NICKEL (II) ION, xylanase Y | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Basis for the Specificity of the Reducing End Xylose-releasing Exo-oligoxylanase from Bacillus halodurans C-125
J.Biol.Chem., 280, 2005
|
|
1T4S
 
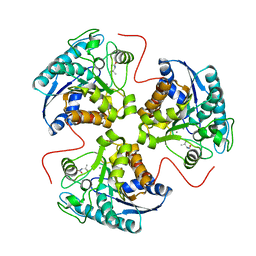 | | arginase-L-valine complex | | Descriptor: | Arginase 1, MANGANESE (II) ION, VALINE | | Authors: | Cama, E, Pethe, S, Boucher, J.-L, Shoufa, H, Emig, F.A, Ash, D.E, Viola, R.E, Mansuy, D, Christianson, D.W. | | Deposit date: | 2004-04-30 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibitor coordination interactions in the binuclear manganese cluster of arginase
Biochemistry, 43, 2004
|
|
1WU6
 
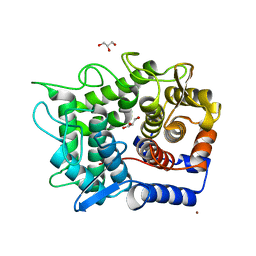 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase E70A mutant complexed with xylobiose | | Descriptor: | GLYCEROL, NICKEL (II) ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis for the Specificity of the Reducing End Xylose-releasing Exo-oligoxylanase from Bacillus halodurans C-125
J.Biol.Chem., 280, 2005
|
|
1UGP
 
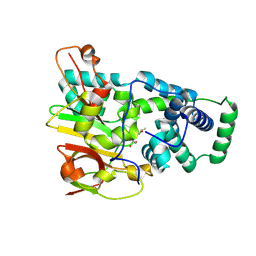 | | Crystal structure of Co-type nitrile hydratase complexed with n-butyric acid | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta, ... | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
1UGS
 
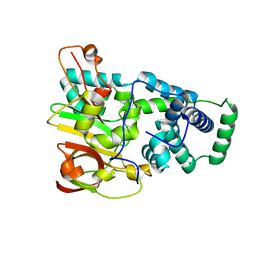 | | Crystal structure of aY114T mutant of Co-type nitrile hydratase | | Descriptor: | COBALT (II) ION, Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
1UGR
 
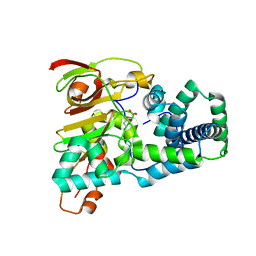 | | Crystal structure of aT109S mutant of Co-type nitrile hydratase | | Descriptor: | COBALT (II) ION, Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
1T4R
 
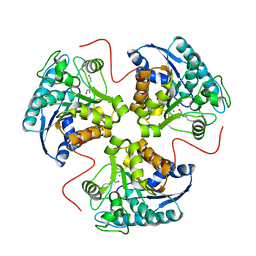 | | arginase-descarboxy-nor-NOHA complex | | Descriptor: | 3-{[(E)-AMINO(HYDROXYIMINO)METHYL]AMINO}PROPAN-1-AMINIUM, Arginase 1, MANGANESE (II) ION | | Authors: | Cama, E, Pethe, S, Boucher, J.-L, Shoufa, H, Emig, F.A, Ash, D.E, Viola, R.E, Mansuy, D, Christianson, D.W. | | Deposit date: | 2004-04-30 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Inhibitor coordination interactions in the binuclear manganese cluster of arginase
Biochemistry, 43, 2004
|
|
1V7V
 
 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase | | Descriptor: | CALCIUM ION, chitobiose phosphorylase | | Authors: | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
